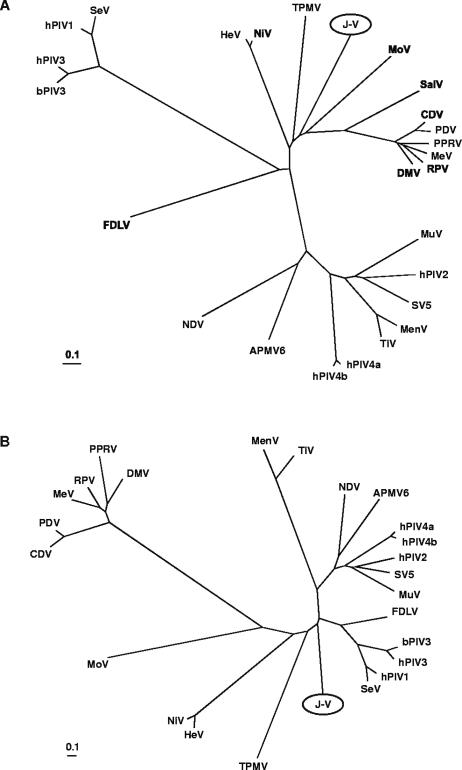
FIG. 7.
Unrooted phylogenetic trees based on complete nucleoprotein (A) and attachment protein (B) sequences of selected viruses within the subfamily Paramyxovirinae. The trees were generated from ClustalW (accurate) protein alignments using distance matrix programs (Protdist and Neighbor) within the PHYLIP software package and were drawn in TreeView. Branch lengths represent relative genetic distances. See Materials and Methods for database accession numbers.