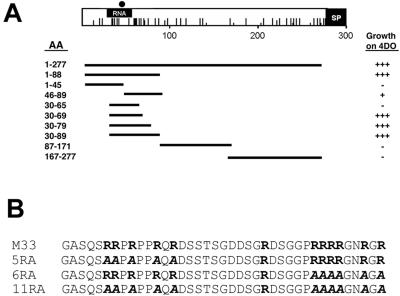
FIG. 5.
Mapping of the p32-binding region of capsid. (A) The p32-binding site is located in the amino-terminal region of capsid. AH109 yeast cells were transformed with pGAD-p32, and plasmids encoding the entire capsid (amino acid residues 1 to 277) or portions thereof were fused to the GAL4-binding domain. The capsid constructs are named according to the amino acid residues (AA) in capsid that they encode. For example, 1-45 is the coding region for amino acid residues 1 to 45 of capsid fused to the GAL4-binding domain. Solid lines indicate the regions of capsid encoded by the constructs. Transformants were plated onto medium lacking tryptophan, leucine, histidine, and adenine, and growth was scored visually in relation to positive and negative controls as follows: −, no growth; +, weak growth; +++, strong growth. The RNA-binding region (RNA), the E2 signal peptide (SP), positively charged residues (vertical lines), and serine 46 (solid circle) are indicated on the capsid schematic. (B) The p32-binding region of wild-type capsid (amino acid residues 30 to 69) contains two clusters of arginine residues (bold letters). Arginine residues in each cluster were changed to alanine residues (bold letters) as indicated, and the resulting mutants were designated 5RA, 6RA, and 11RA.