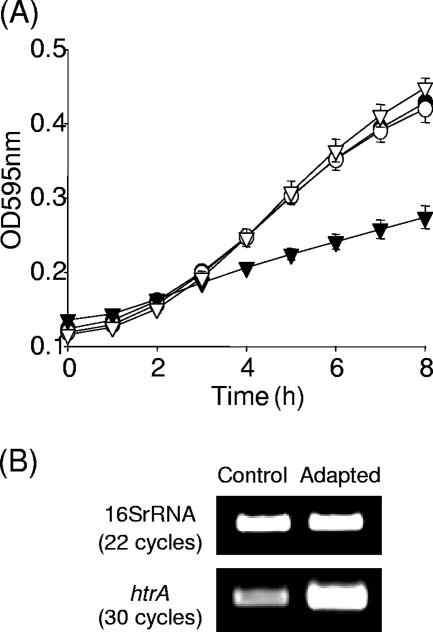
FIG. 3.
(A) Growth of wild-type EGDe (•), revertant (○), htrA mutant (▾), and HtrASS* (▿) under sublethal acid conditions (pH 5, HCl). Error bars represent standard deviations of triplicate experiments. (B) Transcriptional analysis of htrA by RT-PCR. Total RNA was isolated from exponential-phase cultures of EGDe grown in pH 7 BHI (Control) and exposed to pH 5 for 30 min (Adapted). RNA was converted to cDNA, and PCRs were performed with htrA specific primers.