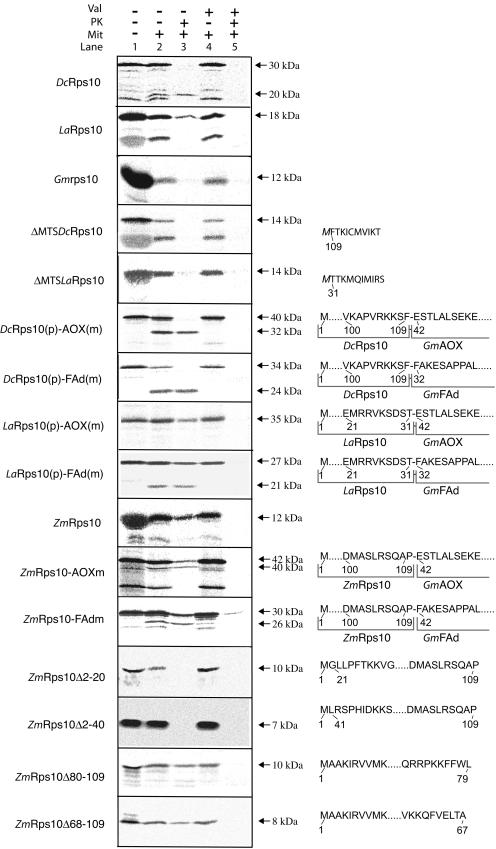
Figure 2.
Import of Rps10 proteins into mitochondria. In vitro radiolabeled Rps10 proteins from carrot, lettuce, maize, and soybean were tested for import ability into purified mitochondria. Import of the various Rps10 wild-type proteins, carrot, and lettuce that have the N-terminal extension removed (ΔMTS) and the carrot and lettuce N-terminal extension placed in front of the alternative oxidase mature protein [DcRps10(p)-AOX(m) and LaRps10(p)-AOX(m)] and in front of the FAd subunit of mitochondrial ATP synthase [DcRps10(p)-FAd(m) and LaRps10(p)-FAd(m)]. The first 10 amino acids are shown for carrot and lettuce where the N-terminal extension has been removed; the M in italics indicates that this residue was added to provide a start codon. The N-terminal extension of carrot and lettuce place in front of the alternative oxidase and FAd subunit are shown with the 10 amino acids from each protein that border the junction point. Numbering is as from Figure 1, with dots indicating any number of amino acids. B, Localization of the mitochondrial targeting information in maize Rps10. The maize Rps10 protein was fused to the mature alternative oxidase and FAd subunit of mitochondrial ATP synthase to test if it could support import of other proteins [ZmRps10-AOX(m) and ZmRps10-FAd(m)]. Deletions were made from the N and C termini to determine the location of the mitochondrial targeting signal in the maize Rps10 protein. In the case of the chimeric proteins, the 10 amino acids boarding the junction are shown. For the deletions, the 10 amino acids nearest the deletion are shown. The numbering of the deletions is based on amino acids number as in Figure 1.