Abstract
Cyclic nucleotide PDE4 (phosphodiesterase 4) inhibitors are being developed as potent anti-inflammatory drugs for use in chronic lung diseases, but the complexity of the PDE4 family has hampered this process. The four genes comprising the PDE4 family, PDE4A, PDE4B, PDE4C and PDE4D, are all expressed as multiple splice variants. The most widely used criterion to identify PDE4 variants expressed endogenously is their migration on SDS/PAGE. However, when a PDE4D3-selective antibody was used for immunoprecipitation, the pattern of expression obtained did not confirm the expression predicted by SDS/PAGE. This observation, together with the recent discovery of additional PDE4D transcripts, prompted us to re-evaluate the pattern of expression of these variants. The nine rat PDE4D splice variants, PDE4D1 to PDE4D9, were cloned, their electrophoretic properties compared, and their in vivo mRNA and protein levels determined. Using this approach, we found that the pattern of distribution of the PDE4D splicing variants is more complex than previously reported. Multiple variants co-migrate in single immunoreactive bands, and variant-selective antibodies were necessary to discriminate between splice variants. Tissues that were thought to express only PDE4D3, express three closely related proteins, with PDE4D8 and PDE4D9 as the predominantly expressed forms. In addition, activation of cAMP signalling produces phosphorylation and activation of variants other than PDE4D3, and expression of PDE4D mRNA does not always correlate with the pattern of protein expression. As PDE4 inhibitors have different affinities for distinct PDE4D splicing variants, our results indicate that a better definition of the pattern of PDE4 expression is required for target validation.
Keywords: cAMP phosphodiesterase, FRTL5 thyroid cell, phosphodiesterase 4 splice form (PDE4 splice form), protein kinase A activation, SDS/PAGE migration, thyroid-stimulating hormone (TSH)
Abbreviations: AMV, avian myeloblastosis virus; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PDE, phosphodiesterase; PKA, cAMP-dependent protein kinase/protein kinase A; poly(A)+, polyadenylated; RT-PCR, reverse transcriptase PCR; TSH, thyroid-stimulating hormone; UCR, upstream conserved region
INTRODUCTION
Degradation of the second messenger cAMP by PDEs (phosphodiesterases) plays an important role in the control of cyclic nucleotide signalling. In addition to terminating cAMP transients, PDEs allow for specificity of signalling by fine-tuning of cAMP levels and the generation of cAMP microdomains [1–4]. These concepts have prompted a re-evaluation of the properties of the PDE forms which hydrolyse cAMP and their expression in any given cell. The recognition of PDEs as promising drug targets has further heightened the interest in these enzymes [5–8].
Type 4 PDEs, the mammalian orthologues of the Drosophila dunce PDE, comprise a distinct family of proteins encoded by four genes [9,10]. These are the major forms involved in cAMP hydrolysis and their expression is essential for many physiological processes, as determined by the use of PDE4-selective inhibitors and by ablation of these genes [11,12].
One property of the dunce gene and its mammalian orthologues is the complex arrangement of the transcriptional units and the splicing variants produced. The Drosophila dunce gene encompasses 150 kb of DNA and more than 15 transcriptional units [13]. A similar complexity was observed in rat and human PDE4 genes [14,15]. The PDE4 loci have multiple transcriptional units that code for two subclasses of variants, termed short and long forms [16]. These forms are distinguished by the presence or absence of two conserved N-terminal domains called UCR1 and UCR2 (upstream conserved regions 1 and 2; [15]). The UCR1/2-module mediates the regulation of catalytic activity triggered by PKA (cAMP-dependent protein kinase) and ERK2 (extracellular-signal-regulated kinase 2) phosphorylation [17–19] and phosphatidic-acid binding [20] and is involved in dimerization [21,22].
The unique N-terminal regions of individual splice variants were shown in many cases to mediate protein–protein or protein–membrane interactions. In addition, individual splice variants are differently regulated by other signalling pathways through post-translational modifications, providing the cell with an array of PDEs that can be targeted to distinct subcellular compartments and associated with distinct signalling pathways [9,10]. The current hypothesis is that each splice variant serves a particular physiological function and an array of diverse promoters enables the expression of distinct subsets of cAMP-hydrolytic activities to meet the demands of any given cell. In support of this concept, effects of PDE4 inhibitors and knock-out mice have been shown to be mediated by single PDE4 variants. The lipopolysaccharide-mediated tumour necrosis factor α response in leucocytes, for example, depends on the expression of the splice variant PDE4B2 [12]. Also, PDE4D long forms have been suggested to mediate the known emetic effects of PDE4 inhibitors [23], and the splice variant PDE4D7 has been associated with a risk haplotype for stroke [24].
The most common criterion used to identify PDE4D forms expressed in any given cell is their migration in SDS/PAGE. Using this criterion, the expression of the short forms, PDE4D1 and PDE4D2, and the long forms, PDE4D3–PDE4D5, has been reported [25–27]. To further characterize the PDE4D isoforms expressed endogenously, we generated antibodies against the N-termini of individual PDE4D variants. When using these antibodies for immunoprecipitation, we observed that only a fraction of discrete immunoreactive bands could be depleted from cell extracts. This observation, together with the recent discovery of additional PDE4D splice variants [24,28], prompted us to reconsider the expression pattern of these proteins. Using deposited DNA sequences, we cloned the rat PDE4D splice variants 1 to 9, compared their enzymatic and regulatory properties, re-assessed their mRNA and protein expression levels in vivo, and established the necessary tools to investigate the function and regulation of distinct variants.
EXPERIMENTAL
PCR primers
The oligonucleotides used for PCR amplification and Southernblot hybridization are numbered consecutively, restriction enzyme sites are underlined, and ATG start codons are shown in italics within the nucleotide sequences: P1, 5′-TATGAAGGAGCAGCCCTCATG-3′; P2, 5′-CCAGGACATCTTCCTGCTCTG-3′; P3, 5′-CAGAAGGCATTCCTGGATATG-3′; P4, 5′-TGGCCAGTTTCTGGTAGGCCTC-3′; P5, 5′-GAGCTACCCGTGGTCGCTAC-3′; P6, 5′-AACCACGTTGCCCTGGATTGT-3′; P7, 5′-CACATTTTAGAACTTGCTGTCAC-3′; P8, 5′-ACTACTCAAAACCGCACCATGG-3′; P9, 5′-GCCACAAGTGCCTCTTGCAGC-3′; P10, 5′-TCCAGACACCAGTCCAGCTCCTCCA-3′; P11, 5′-ATGAGCATTATTATGAAGCCG-3′; P12, 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′; P13, 5′-CATGTAGGCCATGAGGTCCACCAC-3′; P14, 5′-TGGCCAGTTTCTGGTAGGCCTC-3′; P15, 5′-CGATGTGGACAATGGCAC-3′; P16, 5′-TGGAGACTGGTCACCGGC-3′; P17, 5′-ATGGCTCAGCAGACGACAAGC-3′; P18, 5′-CTATCAAAAATGCCTGAAGCAA-3′; P19, 5′-CTTAGCCCTCGCTTTGCCAATG-3′; P20, 5′-TGAGGACAACGGAGGCAGTTTG-3′; P21, 5′-CTACATGGTCTACATGTTCCAG-3′; P22; 5′-GGGGCGAAGCTTATGGCTCAGCAGACGACAAGC-3′; P23, 5′-GCGAAGCTTCGAATTCATGGAGAGGAACACTTGCG-3′; P24, 5′-GCGAAGCTTCGAATTCATGGCTTTCGTTTGGGATCCTC-3′; P25, 5′-GGGGCGAAGCTTATGAGCATTATTATGAAGCCG-3′; P26, 5′-GCGAAGCTTCGAATTCATGGCCTCCAACAAGTTTAAAAGG-3′; P27, 5′-GCGAAGCTTCGAATTCATGCCTGAAGCAAACTATTTATTG-3′.
RNA isolation from rat tissues, RT-PCR (reverse transcriptase PCR) and Southern-blot analysis
Total RNA was isolated from adult rat tissues, including cerebral cortex, cerebellum, spleen, lung, heart, liver, kidney and testis, using TRIzol® reagent in accordance with the manufacturer's instructions (Invitrogen, Carlsbad, CA, U.S.A.), followed by extraction of the poly(A)+ (polyadenylated) mRNA using the Oligotex mRNA isolation kit from Qiagen (Valencia, CA, U.S.A.). Each 500 ng of poly(A)+ mRNA was then reverse transcribed using oligo-dT primers and AMV (avian myeloblastosis virus) reverse transcriptase (Invitrogen). Similar amounts of the resulting first-strand cDNA were used as template for 32 cycles of PCR reactions using the following primer pairs respectively: PDE4D1 and PDE4D2, P1/P2; PDE4D3, P3/P4; PDE4D4, P5/P4; PDE4D5, P6/P4; PDE4D6, P7/P2; PDE4D7, P8/P4; PDE4D8, P9/P10; PDE4D9, P11/P4; GAPDH (glyceraldehyde-3-phosphate dehydrogenase), P12/P13. The expected sizes of amplified fragments are 512 bp, 426 bp, 398 bp, 564 bp, 689 bp, 363 bp, 667 bp, 602 bp and 442 bp for PDE4D1 through PDE4D9 respectively, and 983 bp for GAPDH. All primer pairs are intron–exon spanning, excluding the possibility of artifacts caused by genomic DNA contaminations. The PCR products were subjected to electrophoresis on 1% agarose gels and then transferred on to Biotrans nylon membranes (ICN Pharmaceuticals, Costa Mesa, CA, U.S.A.) for Southern-blot hybridization. To confirm the identity of the PCR products, oligonucleotides specific to the PCR fragments of individual variants were labelled with [γ-32P]ATP by T4 polynucleotide kinase (Roche Diagnostics, Indianapolis, IN, U.S.A.) and used to hybridize the membranes. The oligonucleotide probes used were: PDE4D1 and PDE4D2, P14; PDE4D3 and PDE4D7, P15; PDE4D4, P16; PDE4D5, P17; PDE4D6, P18; PDE4D8, P19; PDE4D9, P20; GAPDH, P21. The membranes were then washed in 2×SSC (300 mM NaCl, 30 mM sodium citrate) containing 0.1% SDS at 50 °C, dried and exposed to X-ray film.
Design of PDE4D expression vectors
PCR was routinely performed using Pwo DNA polymerase (Roche Diagnostics), according to the manufacturer's instructions. The generation of PDE4D1 and PDE4D3 expression vectors has been described previously [29,30]. In brief, the open reading frame of rat PDE4D1 was inserted into the EcoRI restriction enzyme site of the pCMV5 vector and the open reading frame of rat PDE4D3 was cloned between the HindIII and the EcoRI restriction enzyme sites of the pcDNA3 vector (Invitrogen) respectively. The long PDE4D splice variants share a conserved ClaI restriction enzyme site, encoded by their first common exon. We used this internal ClaI site, as well as the HindIII site located upstream of the start codon of the PDE4D3-pcDNA3 vector, to generate expression vectors for other PDE4D long forms. DNA fragments encoding for the 5′-end of rat PDE4D5, PDE4D7, PDE4D8 and PDE4D9 were amplified from various rat tissues (see Figure 5) using primer pairs P22/P4, P23/P4, P24/P4 and P25/P4 respectively. The PCR fragments were then digested with ClaI and HindIII and ligated into the PDE4D3-pcDNA3 vector fragment that had been digested with the same endonucleases. A similar strategy was used to generate expression vectors for the short forms PDE4D2 and PDE4D6, except that a conserved BsmI site encoded by common exon E7 was utilized. DNA fragments were PCR amplified from rat tissues with primer pairs P26/P2 and P27/P2 for PDE4D2 and PDE4D6 respectively. They were then digested with HindIII and BsmI and ligated into the same restriction enzymes sites of the PDE4D3-pcDNA3 vector. An expression vector for PDE4D4 was generated by cloning the entire open reading frame into the BamHI–EcoRI restriction enzyme sites of the pcDNA3 vector. All PCR-generated DNA fragments within final expression vectors were sequenced to exclude the possibility of random mutagenesis.
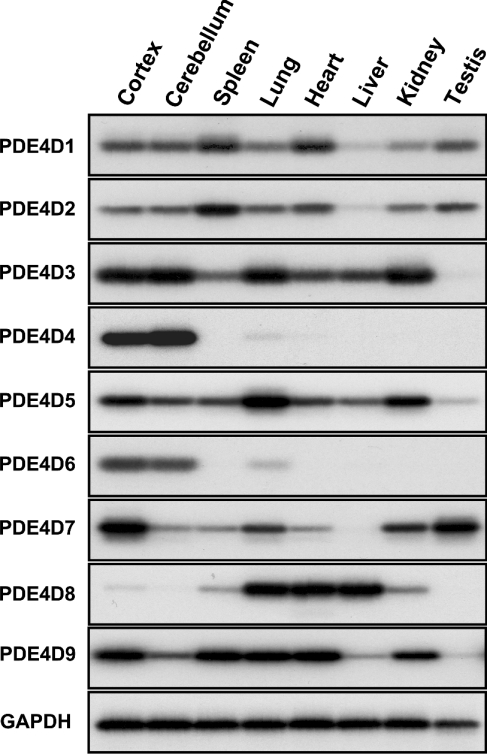
Figure 5. Expression of PDE4D mRNA transcripts in different rat organs.
Transcripts encoding for PDE4D splice variants 1 to 9 were amplified by RT-PCR from poly(A)+ mRNAs isolated from various rat tissues, separated by agarose gel electrophoresis and detected by Southern-blot hybridization, as described in the Experimental section. A representative experiment of the two performed is reported.
Cell culture and transfection
COS7 and HEK-293 cells were cultured at 37 °C and under a 5% CO2 atmosphere in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% fetal calf serum, 30 μg/ml penicillin and 100 μg/ml streptomycin. For recombinant expression of PDE constructs in COS7 cells, cultures were grown on dishes (100-mm diameter) to 50% confluency and were then transfected with recombinant vector DNA using the Effectene™ transfection system (Invitrogen), according to the manufacturer's protocol. After transfection (3 days), cells were routinely harvested in 50 mM Tris/HCl (pH 7.4) containing 1 mM EDTA, 0.2 mM EGTA, 150 mM NaCl, 5 mM 2-mercaptoethanol, 10% glycerol, a protease inhibitor cocktail (Roche Diagnostics; catalogue number, #1836170) and 1 mM AEBSF [4-(2-aminoethyl)benzenesulphonyl fluoride; Roche Diagnostics].
Culture and TSH (thyroid-stimulating hormone) stimulation of FRTL5 cells
FRTL5 cells, a rat thyroid follicular cell line, were cultured in Coon's F12 medium containing 5% calf serum and a mixture of three hormones, including 1 m-unit/ml TSH, 10 μg/ml insulin and 5 μg/ml transferrin, at 37 °C under a 5% CO2 atmosphere. To make the cells quiescent, they were washed twice with HBSS (Hanks balanced salt solution) and then grown in Coon's F12 medium supplemented with 0.1% BSA, but lacking both calf serum and the three hormones the day before the experiment. The next day, cells were treated for 15 min with 10 nM TSH before they were harvested as described above for COS7 and HEK-293 cells.
SDS/PAGE and Western-blot analysis
SDS/PAGE was performed as described by Laemmli [31] using 8% (w/v) polyacrylamide gels. Protein samples were diluted with 4×concentrated Laemmli sample buffer, boiled for 5 min, subjected to electrophoresis and blotted on to Immobilon-P membranes (Millipore, Bedford, MA, U.S.A.). Western-blot analysis was then performed using the polyclonal antibody K116 that recognizes the UCR2C domain of all PDE4 subtypes (α-4PAN) or two monoclonal antibodies which both recognize the PDE4D C-terminus (α-4DPAN; generously given by Pfizer Inc. and Icos Corp.). Immunoreactive bands were detected using the alkaline-phosphatase-conjugated goat anti-mouse/anti-rabbit antibodies and the corresponding detection system from Bio-Rad (Hercules, CA, U.S.A.).
Generation of splice-variant-specific anti-peptide antibodies
To generate splice-variant-specific antisera, peptides encoding the N-terminal regions of PDE4D3 [NH2-MMHVNNFPFRRHSWIC-COOH; aa (amino acids) 1–16], PDE4D4 (NH2-RAGSGEGSDSAGGATLKAPKHLWRHEQHHQYPC-COOH; aa 11–42), PDE4D5 (NH2-EKSKTARKSVSPKLSPVISPRNSPRLLCCOOH; aa 41–67), PDE4D8 (NH2-MAFVWDPLGATVPGPSTRAKSRLRFSKSYC-COOH; aa 1–29) and PDE4D9 (NH2-MSIIMKPRSRSTSSLRTAEAVC-COOH; aa 1–22), with an additional C-terminal cysteine residue where necessary, were synthesized and used to immunize rabbits. The resulting antisera were found to be efficient and variant specific in Western blotting and immunoprecipitation when tested using recombinantly expressed PDE4D variants.
Immunoprecipitation
Protein G–Sepharose beads were washed three times with PBS and then loaded with the respective antibody in a reaction mixture containing 20 μl of Protein G–Sepharose, 100 μl of PBS and 4 μg of the affinity-purified antibody. The samples were incubated in a rotating mixer for 60 min at 4 °C and centrifuged for 2 min at 1000 g. The pelleted beads were washed twice with PBS to remove unbound antibodies, and incubated with the enzyme extracts (500 μg of protein) at 4 °C for 1 h (see Figure 7B) or overnight (all other experiments) in a rotating mixer. The reactions were then centrifuged for 2 min at 1000 g to pellet the immunocomplex. The beads were washed three times by resuspending them in cell lysis buffer and subsequent re-centrifugation. After the last centrifugation step, the beads were resuspended in cell lysis buffer and subjected to PDE assays or the immunoprecipitate was eluted from the sepharose beads for further analysis by SDS/PAGE and Western blotting by boiling the samples for 5 min in 1×Laemmli buffer.
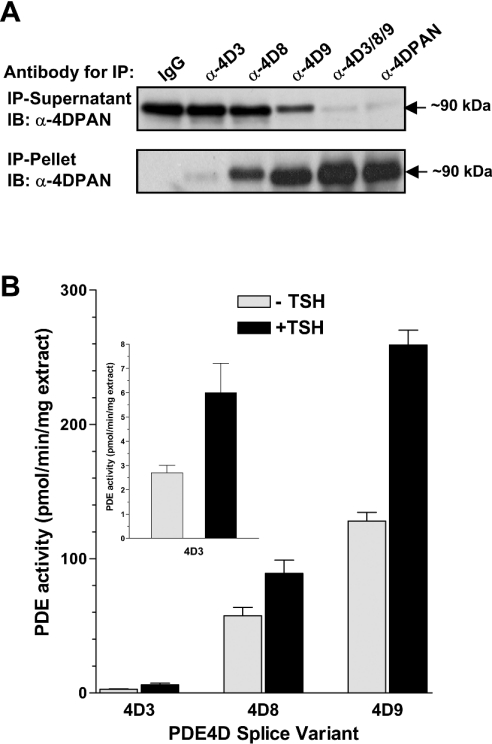
Figure 7. Expression and regulation of PDE4D variants expressed in FRTL5 cells.
(A) Cytosolic extracts prepared from FRTL5 cells were immunoprecipitated with splicevariant-specific antibodies against PDE4D3 (α-4D3), PDE4D8 (α-4D8) and PDE4D9 (α-4D9), a mixture of the three antibodies (α-4D3/8/9), the pan-specific PDE4D antibody M3S1 (α-4DPAN) and IgG as a control. Both the immunoprecipitation (IP) pellets, as well as the depleted immunoprecipitation supernatants, were separated by SDS/PAGE and PDE4D variants were detected by Western blotting using a pan-specific PDE4D antibody (α-4DPAN). (B) Quiescent FRTL5 cells were incubated for 15 min with 10 nM of TSH (+TSH) or without TSH (−TSH) before harvesting the cells. Cytosolic fractions were then subjected to immunoprecipitations using variantspecific PDE4D antibodies. The PDE activities of the immunoprecipitation pellets were determined at 1 μM cAMP. The results shown in the graph are corrected for the unspecific pull-down of PDE activity with purified non-immune IgG, and represent the means±S.E.M. of 4 experiments.
RNA isolation and RT-PCR from FRTL5 cells
Total RNA, isolated from FRTL5 cells using TRIzol® Reagent (Invitrogen), was reverse transcribed using AMV reverse transcriptase (Invitrogen). Similar amounts of the cDNA produced were used as template for PCR reactions using the following primer pairs: PDE4D1 and PDE4D2, P1/P2; PDE4D3, P3/P4; PDE4D4, P5/P4; PDE4D5, P6/P4; PDE4D6, P7/P2; PDE4D7, P8/P4; PDE4D8, P9/P4; PDE4D9, P11/P4. The expected sizes of the amplified fragments were 512 bp, 426 bp, 398 bp, 564 bp, 689 bp, 363 bp, 667 bp, 568 bp and 442 bp for PDE4D1 to PDE4D9 respectively.
PDE assay
PDE activity was measured according to the method of Thompson and Appleman [32], and has been described in detail previously [21,22].
In vitro phosphorylation by PKA
For in vitro phosphorylation experiments, cytosolic supernatants of recombinant PDE constructs expressed in COS7 cells were incubated for 15 min at 30 °C in a reaction mixture containing 2 mM MgCl2, 200 μM ATP, 1 μM microcystin and 1.0 unit of PKA catalytic subunit (Promega, Madison, WI, U.S.A.). Samples were then assayed for PDE activity.
RESULTS
Detection of PDE4D3 in FRTL5 rat thyroid cells
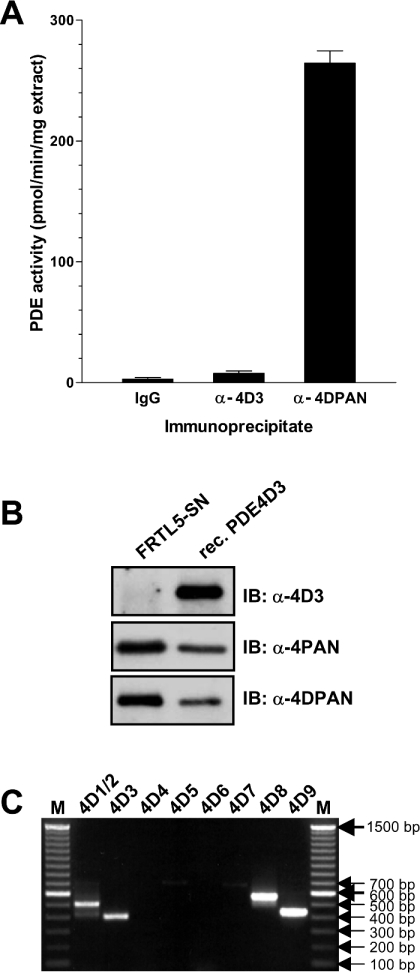
To study the function and regulation of the splice variant PDE4D3, we generated antibodies that selectively recognized this variant. The antiserum raised against an epitope unique to the N-terminus of PDE4D3 detected the recombinant PDE4D3 protein efficiently and selectively in immunoprecipitations and by Western blotting. Surprisingly, however, we found that this antibody could only partially deplete an endogenous PDE4D thought to represent PDE4D3 from cell extracts of the rat thyroid FRTL5 cell line, which had been used extensively in our laboratory [33–35]. In these cells, a major immunoreactive species of PDE4D is detectable in the soluble fraction (Figure 1B; left-hand lane, immunoblot with α-4PAN and α-4DPAN). This band migrates at approx. 90 kDa in SDS/PAGE and was previously identified as PDE4D3 on the basis of co-migration with the corresponding recombinant protein. However, only a minor portion of this band was recognized by the α-4D3 antibody in pull-down assays (Figure 1A), and Western blots with this antibody showed minimal cross-reactivity with this PDE4D species (Figure 1B and see Figure 7A). To resolve this discrepancy, we determined the mRNAs of the PDE4D variants present in FRTL5 cells by RT-PCR. In agreement with our previous reports [33–35], we could detect PDE4D3 message (Figure 1C). However, transcripts for the newly identified PDE4D8 and PDE4D9 [24,28] were also present. Given the possibility that the four recently discovered splice variants PDE4D6 to PDE4D9 (Figure 2) contribute to the overall PDE4D protein in vivo, our aim was to compare the properties of all nine PDE4D splice variants and re-assess their expression patterns and function in vivo using additional form-selective antibodies.
Figure 1. Detection of the PDE4D3 splice variant in the rat thyroid cell line FRTL5.
(A) Cytosolic extracts of FRTL5 were immunoprecipitated using a PDE4D3-specific antibody (α-4D3), the antibody M3S1 that recognizes all PDE4D variants (α-4DPAN) and IgG as a negative control. The graph shows the PDE activity of the immunoprecipitation pellets determined at 1 μM cAMP. (B) A cytosolic extract of FRTL5 cells, as well as recombinant rat PDE4D3, were loaded on to SDS/PAGE and subsequently detected by Western blotting using an antibody that recognizes all PDE4 isoforms (α-4PAN), a pan-specific PDE4D antibody (α-4DPAN) and a PDE4D3 splice-variant-selective antibody (α-4D3). Identical amounts of FRTL5 extract and recombinant PDE were loaded for each of the three Western blots. (C) An agarose gel of PDE4D splice-variant-specific DNA fragments amplified from a FRTL5 cDNA using 35 PCR cycles. The identity of the PCR products was confirmed by restriction digestions (results not shown).
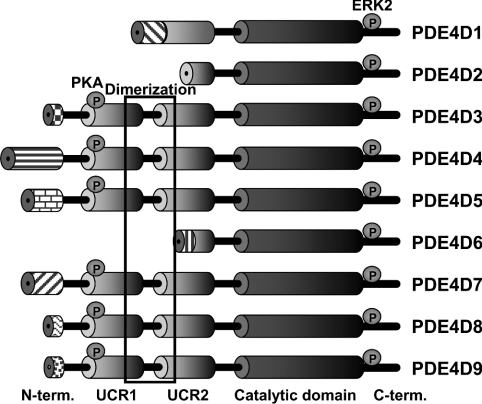
Figure 2. Domain organization of PDE4D protein variants.
Domains are depicted as ‘barrels’ connected by ‘wires’ (putative linker regions). The phosphorylation sites for PKA and ERK2 (extracellular-signal-regulated kinase 2) are shown as circles. The long forms (PDE4D3, PDE4D4, PDE4D5, PDE4D7, PDE4D8 and PDE4D9) and short forms (PDE4D1, PDE4D2 and PDE4D6) are distinguished by the complete or partial presence of the UCR1/2 module respectively.
Properties of recombinant PDE4D splice variants 1 to 9
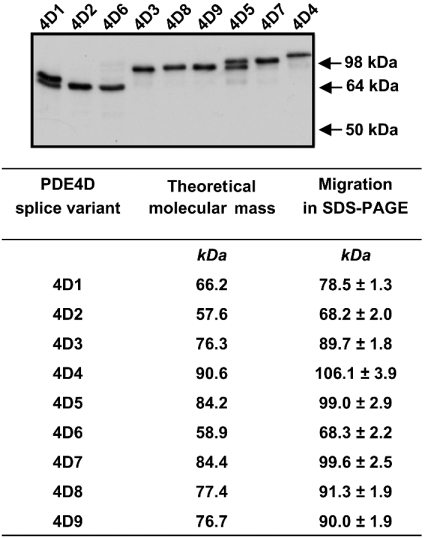
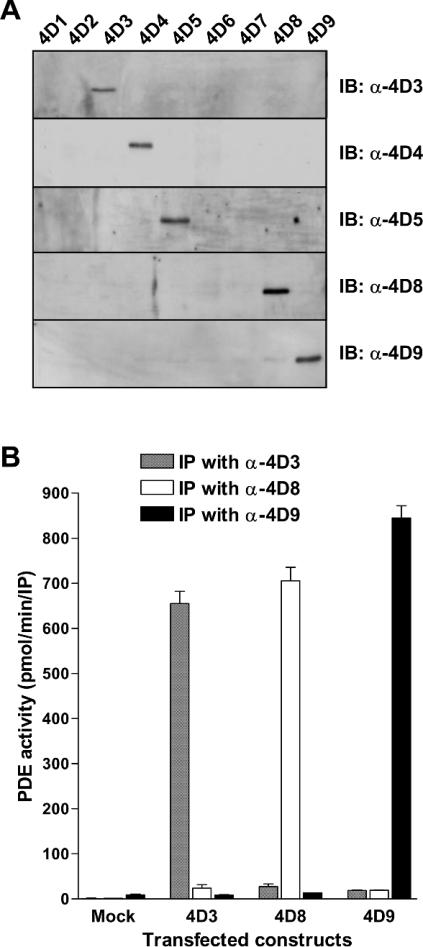
As a first step, we determined the migration of the nine PDE4D variants on SDS/PAGE, as this is the common method to identify PDE4 species expressed endogenously. The open reading frames of variants PDE4D1 to PDE4D9 were amplified by RT-PCR from rat tissues and cloned into mammalian expression vectors, as described in the Experimental section. These expression plasmids were then transfected into COS7 cells. After cell harvest and lysis, soluble extracts of the transfected cells were separated by SDS/PAGE and the recombinant PDE4D proteins were detected by Western blotting using an antibody that recognizes the C-terminus common to all variants (Figure 3). When their migration was compared, three sets of splice variants, PDE4D5 and PDE4D7, the short forms PDE4D2 and PDE4D6, and PDE4D3, PDE4D8, and PDE4D9, exhibited very similar apparent molecular masses in SDS/PAGE (99, 68 and 90 kDa respectively). Thus these PDE4D splice variants were essentially not distinguishable on the basis of their migration in SDS/PAGE. Conversely, antibodies specific for PDE4D3, PDE4D4, PDE4D5, PDE4D8 and PDE4D9 recognized each individual recombinant variant without cross-reactivity in Western blotting and in immunoprecipitations (Figure 4).
Figure 3. Migration of recombinant rat PDE4D splice variants in SDS/PAGE.
Plasmids encoding rat PDE4D splice variants 1 to 9 were transfected in COS7 cells. Cells were harvested after 3 days and cytosolic fractions were subjected to SDS/PAGE and subsequent Western blotting using the K116 antibody that recognizes the UCR2 domain of all PDE4 variants. Similar results were obtained using pan-specific PDE4D antibodies against the PDE4D C-terminus (results not shown). Two PDE4D immunoreactive bands are present in cell extracts over-expressing PDE4D1 and PDE4D5. The slower migrating mRNA band represents the genuine full-length enzyme in both cases. The band below recombinant PDE4D1 is most likely PDE4D2, which may be generated by alternative splicing of the PDE4D1. The PDE4D-immunoreactive protein detected below full-length PDE4D5 may be generated by proteolytic degradation of PDE4D5 or by use of an alternative translation start site (Met71 of the PDE4D5 amino acid sequence).
Figure 4. Detection of recombinant PDE4D with splice-variant-specific antibodies.
(A) Recombinant PDE4D splice variants 1 to 9 were loaded on to SDS/PAGE (6% gels), separated by electrophoresis and blotted on to Immobilon-P membranes. Antibodies raised against the N-terminal domains unique to PDE4D3 (α-4D3), PDE4D4 (α-4D4), PDE4D5 (α-4D5), PDE4D8 (α-4D8) and PDE4D9 (α-4D9) were then used to detect the respective variants by immunoblotting (IB). All panels depict a molecular mass range of approx. 60–180 kDa. (B) COS7 cells were transfected with plasmids encoding PDE4D3, PDE4D8 and PDE4D9 and with the empty pcDNA3-vector as a control (Mock). Cells were harvested after 3 days and each 30 μl of soluble extracts was used for immunoprecipitation (IP) with antibodies specific for PDE4D3 (α-4D3), PDE4D8 (α-4D8) and PDE4D9 (α-4D9), followed by the determination of PDE activity in the immunoprecipitation pellets.
Tissue distribution of PDE4D transcripts in rat
To determine the expression pattern of PDE4D splice variants in vivo, we amplified mRNA transcripts for PDE4D1 to PDE4D9 from various rat organs by RT-PCR and determined their signal intensities after Southern-blot hybridization (Figure 5). The nine PDE4D mRNAs showed profound differences in their tissue distribution. In general, PDE4D4 and PDE4D6 were the mRNAs with the highest selectivity of expression, being abundant in the brain, but not detected in significant amounts in other tissues. PDE4D7 and PDE4D8 were found in a restricted number of tissues and were expressed at highest levels in cortex, testis and kidney (PDE4D7) and in lung, heart and liver (PDE4D8) respectively. Finally, PDE4D1, PDE4D2, PDE4D3, PDE4D5 and PDE4D9 were all widely distributed and present to some extent in most tissues.
The mRNA encoding PDE4D2 is generated by splicing of the PDE4D1 primary transcript. The expression pattern of PDE4D1 and PDE4D2 in rat is almost identical. This may indicate that splicing of PDE4D1 into PDE4D2 transcripts is not regulated.
Identification of endogenous PDE4D proteins
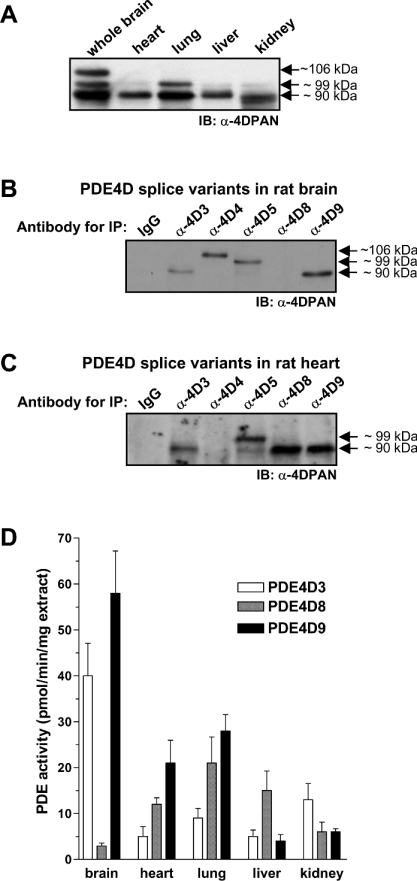
To determine the distribution of PDE4D variants at the protein level, we separated soluble extracts from five rat tissues on SDS/PAGE gels followed by Western blotting with pan-PDE4D-specific antibodies. Three immunoreactive bands of about 90, 99 and 106 kDa were detected (Figure 6A). The brain contains substantial amounts of all three bands, whereas heart, lung, liver, and kidney lack the 106 kDa band, but contain the 90 and 99 kDa proteins. Immunoreactive bands representative of short PDE4D variants were not detected in any of the five tissues (results not shown).
Figure 6. Tissue distributions of PDE4D proteins.
(A) Detergent extracts of organs taken from 1-month-old Sprague–Dawley rats prepared using 1% Nonidet P40 were separated by SDS/PAGE and PDE4D proteins were detected by Western blotting using the pan-specific PDE4D antibody (α-4DPAN). Immunoreactive bands for the short splice variants were not detected in any of the tissues. A representative of three experiments performed is shown. IB, immunoblot. (B, C) Each 500 μg of the soluble extracts prepared from rat whole brain and heart using 1% Nonidet P40 was applied to pull-down assays using variant-specific antibodies for PDE4D3, 4, 5, 8 and 9, followed by the detection of the immunoprecipitated PDE4D variants using a pan-specific PDE4D selective antibody in Western blots. IP, immunoprecipitation. (D) Each 500 μg of the soluble extracts prepared from different rat organs using 1% Nonidet P40 was applied to immunoprecipitations with variant-specific antibodies for PDE4D3, 4, 5, 8 and 9. The immunoprecipitated PDE activities are expressed per mg of protein extract used as input for the immunoprecipitation. The results are expressed as the means±S.E.M. of 4 experiments.
To distinguish endogenous PDE4D splice variants with similar electrophoretic mobility, we generated additional variant-specific antibodies against PDE4D4, PDE4D5, PDE4D8 and PDE4D9 (Figure 4). As shown in Figures 6(B) to 6(D), in addition to PDE4D3, 4 and 5, the splice variants PDE4D8 and 9 are expressed in substantial amounts in the five rat organs studied. PDE4D8 and 9, not PDE4D3, represent the majority of the 90 kDa PDE4D immunoreactive band in almost all tissues (Figure 6D). A similar heterogeneity might be present for the 99-kDa PDE4D-immunoreactive band. When analysing brain extracts that had been depleted of PDE4D5, we found that a band of approx. 99 kDa was still present in Western blots with pan-specific PDE4D antibodies (approx. 20% immunoreactivity remained), but not in blots detected with a PDE4D5-specific antibody (results not shown). The remaining immunoreactivity is likely to be contributed, at least partially, by PDE4D7 as mRNA, for this splice variant is present in the brain (see Figure 5) and its migration in SDS/PAGE is similar to PDE4D5 (see Figure 3).
PDE4D variants expressed in FRTL5 and HEK-293 cells
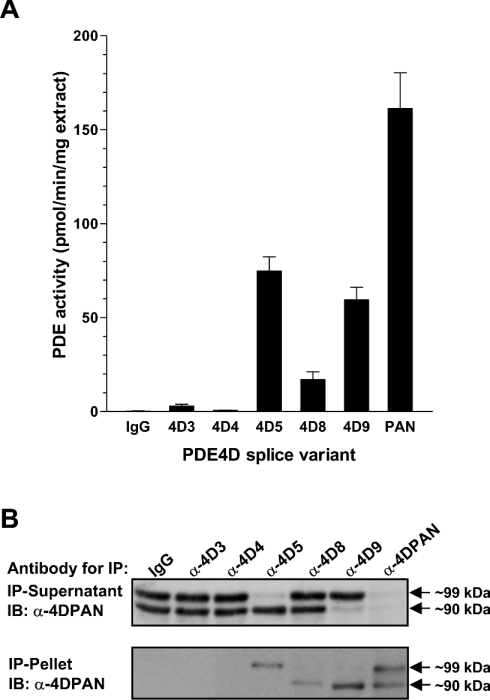
We then used the variant-specific PDE4D antibodies to re-evaluate the PDE4D proteins expressed in the FRTL5 cell line. As shown in Figure 7(A), the PDE4D in FRTL5 is mainly contributed by splice variants PDE4D8 and 9. Treatment of quiescent FRTL5 cells with TSH leads to a rapid phosphorylation and activation of an approx. 90-kDA PDE4D variant, as reported previously [33,34]. As shown in Figure 7(B), PDE4D9, not PDE4D3, is the main form involved in this PKA-mediated feedback regulation of TSH-receptor signalling, as PDE4D9 is the major form expressed and is activated upon TSH treatment to a higher extent than PDE4D8.
The splice variant-selective antibodies against PDE4D3, 5, 8 and 9 cross-react with mouse and human PDE4D. This allowed us to analyse the expression of PDE4D proteins in other mammalian cell lines, such as HEK-293 (Figure 8), COS7 and mouse fibroblast cells (results not shown). In a manner similar to that found in FRTL5 cells we observed that immunoreactive bands previously thought to represent exclusively PDE4D3 or PDE4D5 were only partially depleted by antibodies specific to these variants in Western blots and pull-down assays, and PDE4D9 was again the major 90-kDa PDE4D species expressed.
Figure 8. Expression of PDE4D variants in HEK-293 cells.
(A) Cytosolic extracts prepared from HEK-293 cells were subjected to immunoprecipitation with antibodies specifically recognizing PDE4D3, PDE4D4, PDE4D5, PDE4D8 and PDE4D9, as well as the pan-specific PDE4D antibody M3S1 (PAN) and non-immune IgG as a control. The PDE activities of the immunoprecipitation pellets were then determined at 1 μM cAMP. The results represent the means±S.E.M. of 3 experiments. (B) Cytosolic extracts prepared from HEK-293 cells were immunoprecipitated with several splice-form-specific antibodies, as well as the panspecific PDE4D antibody M3S1 (α-4DPAN) and IgG as a control. Both the immunoprecipitation pellet, as well as the depleted immunoprecipitation supernatant, were separated by SDS/PAGE and PDE4D variants were detected by Western blotting using a pan-specific PDE4D antibody.
PKA-mediated activation of PDE4D splice variants
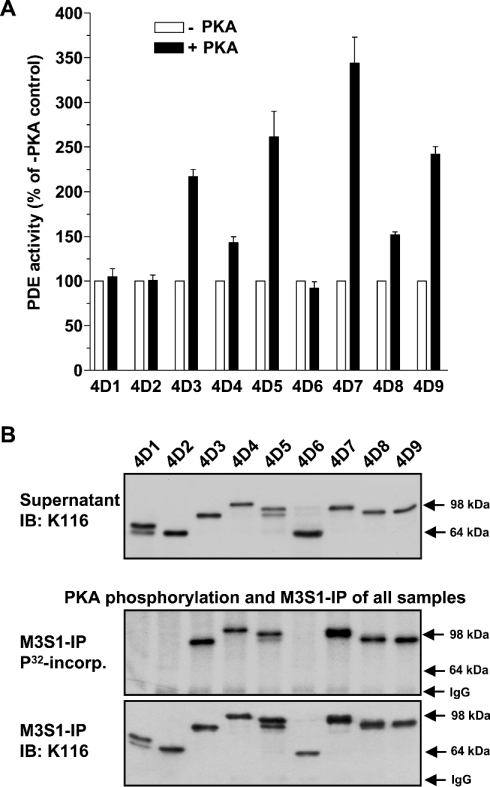
PDE4 long forms share a conserved PKA phosphorylation site within their UCR1 domain. To assess the efficiency of PKA-mediated phosphorylation and regulation of PDE4D splice variants, in vitro phosphorylation assays were performed. As shown in Figure 9(A), all PDE4 long forms were activated, although to different extents, by PKA phosphorylation. Conversely, short PDE4D variants were not phosphorylated or activated by PKA phosphorylation (Figures 9A and 9B).
Figure 9. PKA-mediated activation of PDE4D splice variants.
(A) Cytosolic supernatants of rat PDE4D splice variants 1 to 9 expressed in COS7 cells were incubated for 15 min in an in vitro phosphorylation assay in the presence (+PKA) or absence (−PKA) of 1.0 unit of PKA catalytic subunit prior to the measurement of PDE activity. (B) Cytosolic fractions of COS7 cells over-expressing rat PDE4D splice variants 1 to 9 (upper panel) were incubated for 15 min in an in vitro phosphorylation assay containing 1.0 unit of the catalytic subunit of PKA, 2 μCi of [γ-32P]ATP and 2 mM MgCl2, after which the PDE4D proteins were immunoprecipitated using the anti-PDE4D antibody M3S1 coupled to Protein G. The resulting immunoprecipitates were then analysed for 32P incorporation (middle panel) and PDE4D protein (lower panel) by SDS/PAGE and Western blotting.
DISCUSSION
The four genes comprising the PDE4 family are expressed as more than twenty splice variants, each exhibiting unique properties. Different organs, tissues or cells utilize an array of diverse promoters to express a unique pattern of PDE variants according to their specific requirements, which are still largely unknown. All tissues contain at least one, but often a multitude of splice variants, providing the cells with an array of molecules with specialized functions. PDE4D4, 6, 7 and 8 are selectively transcribed in a restricted number of tissues, whereas PDE4D1, 2, 3, 5 and 9 are widely distributed and are present in most rat tissues (Figure 5). Their tissue-specific expression patterns are retained to some extent across species, which may indicate a conservation of function [28].
Although mRNA for the short forms PDE4D1, 2 and 6 was detected in several tissues (Figure 5) and cell lines (Figure 1C), protein for PDE4D short forms could not be detected by Western blotting using several different antibodies (results not shown). This discrepancy may be explained by the regulation of protein expression of the short forms at the level of translation and/or protein degradation [36]. Also, the message for the short forms may simply be expressed at lower levels compared with the long forms. The differential expression of long- and short-form protein appears in tune with the distinct mechanisms known to regulate their activity. The enzyme activity of PDE4 long forms is usually controlled by post-translational modifications, ligand binding and protein–protein interactions, whereas their protein levels in the cell are relatively stable or change slowly [18,20,34,37]. This concept is consistent with the predominant presence of the long-form protein in all cells and tissues analysed in Figures 6–8. Conversely, the activity of short PDE4 variants (such as PDE4D1, PDE4D2 or PDE4B2) is often determined by major changes of transcription/translation levels in response to extracellular signals (>100-fold inductions of mRNA and protein have been reported) [12,29,33]. Thus PDE4 short forms are expressed in a spatially and temporally restricted manner in response to elevated cAMP levels.
The migration in SDS/PAGE has been used to distinguish splice variants PDE4D1 to PDE4D5 when expressed endogenously [25–27]. However, with the recent discovery of four new PDE4D variants (PDE4D6–PDE4D9; [24,28]), it appears that this method is in fact inaccurate, as several sets of splice variants have similar molecular masses and show comparable SDS/PAGE migration (Figure 3). Changes in the electrophoretic behaviour of splice variants upon post-translational modification, such as the phosphorylation of PDE4 long forms by PKA, make the interpretation of SDS/PAGEs and Western blots even more complicated. Considering the wide distribution of mRNA for the new splice variants (Figure 5) and their prevalent contribution to the total PDE4D activity in the cell lines and tissues analysed in this study (Figures 6–8), it is clear that the pattern of tissue distribution of PDE4D variants is more complex than previously thought and should be re-evaluated using alternative methods.
The amount of PDE activity immunoprecipitated for each long splice variant correlated very well, in general, with the distribution of its respective mRNA in the various tissues shown in Figure 5. Using splice-variant-specific antisera to unveil the distinct expression and regulation of PDE4D variants, we show that the major PDE4D species in FRTL5 cells (representing >70% of the total PDE activity), previously thought to be PDE4D3, is instead mainly contributed by PDE4D8 and PDE4D9 (Figure 7). Thus, unlike our previous conclusion, the PKA-mediated PDE4D activation, an important negative feedback regulation of thyroid-receptor activation in these cells [17,34], is mainly mediated by PDE4D9 and to a limited extent by PDE4D8. Similarly, other strategies, including the definition of protein–protein interactions using endogenous proteins, should not be based solely on the migration of a PDE4D variant in SDS/PAGE. Form-selective antibodies need to be used, particularly if interacting domains are present at the N-terminus of a PDE4. More conclusive methods to identify endogenous protein should be considered for all other PDEs as well, because most PDE genes are expressed as several splice variants.
Phosphorylation and activation of PDE4 by PKA constitutes a critical event in the short-term feedback regulation of cAMP transients. Sette and Conti [17] first showed that a long PDE4 form, PDE4D3, is activated by PKA, and identified a serine residue in UCR1 as the phosphorylation site that mediates this effect. Phosphorylation of this residue, which is conserved in all long forms, was subsequently shown to also mediate PKA-dependent activation of the long forms of PDE4A, PDE4B and PDE4C [38,39]. In agreement with the idea that PKA-mediated activation is a conserved regulatory mechanism of PDE4, all long PDE4D splice variants were found to be activated by PKA in the present study (Figure 9). However, the extent of activation varies widely ranging from 40% in PDE4D4 to 350% in PDE4D7. It remains to be determined if these differences also exist in vivo, as over-expression artifacts that might have an impact on the level of PKA-dependent activation cannot be ruled out. The short PDE4D splice variants, which lack the conserved UCR1 phosphorylation site and are not activated by PKA, also lack alternative PKA-phosphorylation sites, as shown by 32P-incorporation experiments.
In summary, the nine splice variants encoded by the PDE4D locus are widely distributed in rat, and most tissues contain a subset of several different PDE4D splice forms. As some splice variants show similar migration in SDS/PAGE, this criterion cannot be used to identify endogenous PDE4D species using nonselective PDE4D antibodies, and alternative means, such as splice variant-specific antibodies, must be sought to unequivocally determine the pattern of expression and the physiological function of distinct splice variants. In addition, mRNA expression of the short forms was not always associated with detectable accumulation of the corresponding proteins.
Acknowledgments
We are indebted to Caren Spencer for editorial work on the manuscript prior to submission. This work was supported by National Institutes of Health grant HD20788 (M.C.) and a fellowship grant from the German Academic Exchange Service (W.R.).
References
- 1.Jurevicius J., Fischmeister R. cAMP compartmentation is responsible for a local activation of cardiac Ca2+ channels by β-adrenergic agonists. Proc. Natl. Acad. Sci. U.S.A. 1996;93:295–299. doi: 10.1073/pnas.93.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rich T. C., Fagan K. A., Tse T. E., Schaack J., Cooper D. M., Karpen J. W. A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13049–13054. doi: 10.1073/pnas.221381398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaccolo M., Magalhaes P., Pozzan T. Compartmentalisation of cAMP and Ca2+ signals. Curr. Opin. Cell Biol. 2002;14:160–166. doi: 10.1016/s0955-0674(02)00316-2. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg S. F., Brunton L. L. Compartmentation of G protein-coupled signaling pathways in cardiac myocytes. Annu. Rev. Pharmacol. Toxicol. 2001;41:751–773. doi: 10.1146/annurev.pharmtox.41.1.751. [DOI] [PubMed] [Google Scholar]
- 5.Shakur Y., Holst L. S., Landstrom T. R., Movsesian M., Degerman E., Manganiello V. Regulation and function of the cyclic nucleotide phosphodiesterase (PDE3) gene family. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:241–277. doi: 10.1016/s0079-6603(00)66031-2. [DOI] [PubMed] [Google Scholar]
- 6.Spina D. Phosphodiesterase-4 inhibitors in the treatment of inflammatory lung disease. Drugs. 2003;63:2575–2594. doi: 10.2165/00003495-200363230-00002. [DOI] [PubMed] [Google Scholar]
- 7.O'Donnell J. M., Zhang H. T. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol. Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Corbin J. D., Francis S. H., Webb D. J. Phosphodiesterase type 5 as a pharmacologic target in erectile dysfunction. Urology. 2002;60:4–11. doi: 10.1016/s0090-4295(02)01686-2. [DOI] [PubMed] [Google Scholar]
- 9.Conti M., Richter W., Mehats C., Livera G., Park J. Y., Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J. Biol. Chem. 2003;278:5493–5496. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- 10.Houslay M. D., Adams D. R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S. L., Richard F. J., Kuo W. P., D'Ercole A. J., Conti M. Impaired growth and fertility of cAMP-specific phosphodiesterase PDE4D-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11998–12003. doi: 10.1073/pnas.96.21.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S. L., Conti M. Induction of the cyclic nucleotide phosphodiesterase PDE4B is essential for LPS-activated TNF-α responses. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7628–7633. doi: 10.1073/pnas.122041599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Y. H., Chen C. N., Malone T., Richter L., Beckendorf S. K., Davis R. L. Characterization of the memory gene dunce of Drosophila melanogaster. J. Mol. Biol. 1991;222:553–565. doi: 10.1016/0022-2836(91)90496-s. [DOI] [PubMed] [Google Scholar]
- 14.Swinnen J. V., Joseph D. R., Conti M. Molecular cloning of rat homologues of the Drosophila melanogaster dunce cAMP phosphodiesterase: evidence for a family of genes. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5325–5329. doi: 10.1073/pnas.86.14.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger G., Michaeli T., Martins T., St John T., Steiner B., Rodgers L., Riggs M., Wigler M., Ferguson K. A family of human phosphodiesterases homologous to the dunce learning and memory gene product of Drosophila melanogaster are potential targets for antidepressant drugs. Mol. Cell Biol. 1993;13:6558–6571. doi: 10.1128/mcb.13.10.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conti M., Nemoz G., Sette C., Vicini E. Recent progress in understanding the hormonal regulation of phosphodiesterases. Endocrinol. Rev. 1995;16:370–389. doi: 10.1210/edrv-16-3-370. [DOI] [PubMed] [Google Scholar]
- 17.Sette C., Conti M. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J. Biol. Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann R., Baillie G. S., MacKenzie S. J., Yarwood S. J., Houslay M. D. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 1999;18:893–903. doi: 10.1093/emboj/18.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houslay M. D., Baillie G. S. The role of ERK2 docking and phosphorylation of PDE4 cAMP phosphodiesterase isoforms in mediating cross-talk between the cAMP and ERK signalling pathways. Biochem. Soc. Trans. 2003;31:1186–1190. doi: 10.1042/bst0311186. [DOI] [PubMed] [Google Scholar]
- 20.Grange M., Sette C., Cuomo M., Conti M., Lagarde M., Prigent A. F., Nemoz G. The cAMP-specific phosphodiesterase PDE4D3 is regulated by phosphatidic acid binding. Consequences for cAMP signaling pathway and characterization of a phosphatidic acid binding site. J. Biol. Chem. 2000;275:33379–33387. doi: 10.1074/jbc.M006329200. [DOI] [PubMed] [Google Scholar]
- 21.Richter W., Conti M. Dimerization of the type 4 cAMP-specific phosphodiesterases is mediated by the upstream conserved regions (UCRs) J. Biol. Chem. 2002;277:40212–40221. doi: 10.1074/jbc.M203585200. [DOI] [PubMed] [Google Scholar]
- 22.Richter W., Conti M. The oligomerization state determines regulatory properties and inhibitor sensitivity of type 4 cAMP-specific phosphodiesterases. J. Biol. Chem. 2004;279:30338–30348. doi: 10.1074/jbc.M312687200. [DOI] [PubMed] [Google Scholar]
- 23.Robichaud A., Stamatiou P. B., Jin S. L., Lachance N., MacDonald D., Laliberte F., Liu S., Huang Z., Conti M., Chan C. C. Deletion of phosphodiesterase 4D in mice shortens α2-adrenoceptor-mediated anesthesia, a behavioral correlate of emesis. J. Clin. Invest. 2002;110:1045–1052. doi: 10.1172/JCI15506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gretarsdottir S., Thorleifsson G., Reynisdottir S. T., Manolescu A., Jonsdottir S., Jonsdottir T., Gudmundsdottir T., Bjarnadottir S. M., Einarsson O. B., Gudjonsdottir H. M., et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 25.Iona S., Cuomo M., Bushnik T., Naro F., Sette C., Hess M., Shelton E. R., Conti M. Characterization of the rolipram-sensitive, cyclic AMP-specific phosphodiesterases: identification and differential expression of immunologically distinct forms in the rat brain. Mol. Pharmacol. 1998;53:23–32. doi: 10.1124/mol.53.1.23. [DOI] [PubMed] [Google Scholar]
- 26.Bolger G. B., McCahill A., Huston E., Cheung Y. F., McSorley T., Baillie G. S., Houslay M. D. The unique amino-terminal region of the PDE4D5 cAMP phosphodiesterase isoform confers preferential interaction with β-arrestins. J. Biol. Chem. 2003;278:49230–49238. doi: 10.1074/jbc.M303772200. [DOI] [PubMed] [Google Scholar]
- 27.Liu H., Palmer D., Jimmo S. L., Tilley D. G., Dunkerley H. A., Pang S. C., Maurice D. H. Expression of phosphodiesterase 4D (PDE4D) is regulated by both the cyclic AMP-dependent protein kinase and mitogen-activated protein kinase signaling pathways. A potential mechanism allowing for the coordinated regulation of PDE4D activity and expression in cells. J. Biol. Chem. 2000;275:26615–26624. doi: 10.1074/jbc.M001634200. [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Deng C., Bugaj-Gaweda B., Kwan M., Gunwaldsen C., Leonard C., Xin X., Hu Y., Unterbeck A., De Vivo M. Cloning and characterization of novel PDE4D isoforms PDE4D6 and PDE4D7. Cell. Signal. 2003;15:883–891. doi: 10.1016/s0898-6568(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 29.Sette C., Vicini E., Conti M. The rat PDE3/IVd phosphodiesterase gene codes for multiple proteins differentially activated by cAMP-dependent protein kinase. J. Biol. Chem. 1994;269:18271–18274. [PubMed] [Google Scholar]
- 30.Swinnen J. V., Joseph D. R., Conti M. The mRNA encoding a high-affinity cAMP phosphodiesterase is regulated by hormones and cAMP. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8197–8201. doi: 10.1073/pnas.86.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Thompson W. J., Appleman M. M. Multiple cyclic nucleotide phosphodiesterase activities from rat brain. Biochemistry. 1971;10:311–316. [PubMed] [Google Scholar]
- 33.Jin S. L., Bushnik T., Lan L., Conti M. Subcellular localization of rolipram-sensitive, cAMP-specific phosphodiesterases. Differential targeting and activation of the splicing variants derived from the PDE4D gene. J. Biol. Chem. 1998;273:19672–19678. doi: 10.1074/jbc.273.31.19672. [DOI] [PubMed] [Google Scholar]
- 34.Oki N., Takahashi S. I., Hidaka H., Conti M. Short term feedback regulation of cAMP in FRTL-5 thyroid cells. Role of PDE4D3 phosphodiesterase activation. J. Biol. Chem. 2000;275:10831–10837. doi: 10.1074/jbc.275.15.10831. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi S. I., Nedachi T., Fukushima T., Umesaki K., Ito Y., Hakuno F., Van Wyk J. J., Conti M. Long-term hormonal regulation of the cAMP-specific phosphodiesterases in cultured FRTL-5 thyroid cells. Biochim. Biophys. Acta. 2001;1540:68–81. doi: 10.1016/s0167-4889(01)00119-7. [DOI] [PubMed] [Google Scholar]
- 36.Conti M., Jin S. L. The molecular biology of cyclic nucleotide phosphodiesterases. Prog. Nucleic Acid Res. Mol. Biol. 1999;63:1–38. doi: 10.1016/s0079-6603(08)60718-7. [DOI] [PubMed] [Google Scholar]
- 37.Bolger G. B., Peden A. H., Steele M. R., MacKenzie C., McEwan D. G., Wallace D. A., Huston E., Baillie G. S., Houslay M. D. Attenuation of the activity of the cAMP-specific phosphodiesterase PDE4A5 by interaction with the immunophilin XAP2. J. Biol. Chem. 2003;278:33351–33363. doi: 10.1074/jbc.M303269200. [DOI] [PubMed] [Google Scholar]
- 38.Laliberte F., Liu S., Gorseth E., Bobechko B., Bartlett A., Lario P., Gresser M. J., Huang Z. In vitro PKA phosphorylation-mediated human PDE4A4 activation. FEBS Lett. 2002;512:205–208. doi: 10.1016/s0014-5793(02)02259-7. [DOI] [PubMed] [Google Scholar]
- 39.MacKenzie S. J., Baillie G. S., McPhee I., MacKenzie C., Seamons R., McSorley T., Millen J., Beard M. B., van Heeke G., Houslay M. D. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in upstream conserved region 1 (UCR1) Br. J. Pharmacol. 2002;136:421–433. doi: 10.1038/sj.bjp.0704743. [DOI] [PMC free article] [PubMed] [Google Scholar]