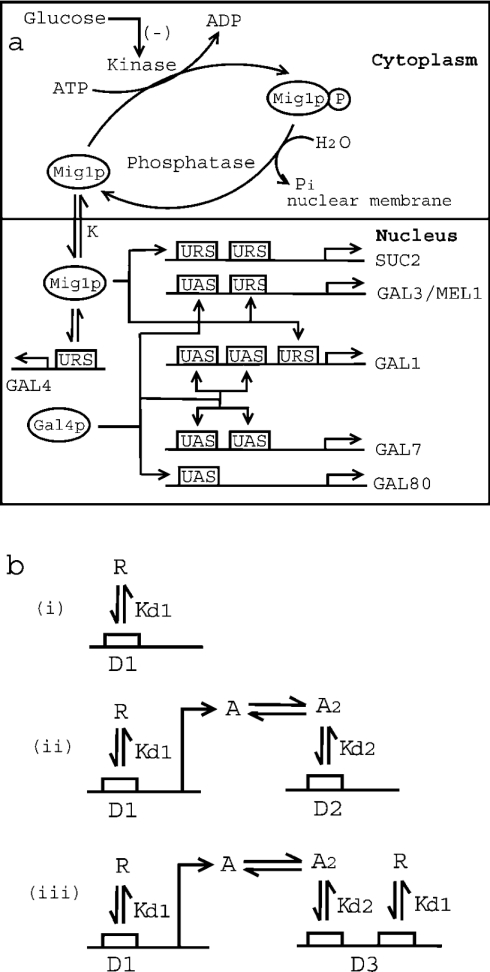
Figure 1. Mechanism of glucose repression in S. cerevisiae.
(a) Schematic representation of Mig1p-mediated glucose repression of SUC2 and GAL genes. Kinase (Snf1) phosphorylates Mig1p to Mig1p-P (phosphorylated product), while phosphatase dephosphorylates Mig1p. Glucose inhibits the activity of Snf1 kinase. Unphosphorylated Mig1p is translocated to the nucleus with a distribution coefficient of K. Furthermore, Mig1p in the nucleus binds to the URS of SUC2, GAL4, GAL1, GAL3 and MEL1, with a dissociation constant Kd1. It may be noted that SUC2 has two binding sites for Mig1p, while GAL4, GAL1, GAL3 and MEL1 have only one binding site. Also, the transcriptional activator, Gal4p, binds to the UAS of GAL3/MEL1 and GAL80 (with one binding site for Gal4p), and GAL1 and GAL7 (with two binding sites for Gal4p). (b) Schematic representation of protein–protein and protein–DNA interactions for three different mechanisms for gene repression. Mechanism of repression in which (i) repressor protein ‘R’ binds to the URS of gene ‘D1’ with a dissociation constant of Kd1, (ii) repressor protein ‘R’ binds to the URS of transcriptional activator gene ‘D1’. The product of gene ‘D1’ is transcriptional activator protein ‘A’, which dimerizes with dissociation constant Kd1 before binding to the UAS of gene ‘D2’ with dissociation constant Kd2. (iii) Both mechanisms (i) and (ii) described above are operational.