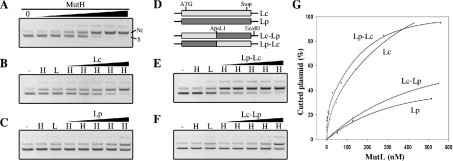
Figure 4. In vitro activation of E. coli MutH endonuclease activity.
(A) Unmethylated d(GATC) plasmid was digested with increasing amounts of E. coli MutH protein (see the Experimental section), separated on 0.6% agarose gel and visualized with ethidium bromide. S, supercoiled plasmid; Nc, nicked circular plasmid. (B, C, E and F) Unmethylated d(GATC) plasmid was incubated with buffer (–), with 70 nM E. coli MutH (H), with 500 nM of the indicated MutL protein (L) or with 70 nM E. coli MutH and increasing amounts of the indicated MutL protein (H/MutL) (see the Experimental section), before separation on 0.6% agarose gel. (D) Schematic representation of E. coli (Lc), P. aeruginosa (Lp) and chimaeric MutL proteins containing the E. coli N-terminal region and P. aeruginosa C-terminal region (Lc–Lp) or vice versa (Lp–Lc). ApaLI and EcoRI indicate positions of the corresponding restriction sites used for the construction of chimaeric mutL genes. (G) Ethidium bromide-stained gels (B, C, E and F) were scanned and the amount of Nc plasmid was plotted as a percentage. The amount of Nc plasmid obtained with MutH alone (70 nM) was defined as 0%. The difference between the total amount of plasmid used and the amount of Nc plasmid obtained with MutH alone is taken as 100%. Negatives of ethidium bromide photographs are shown.