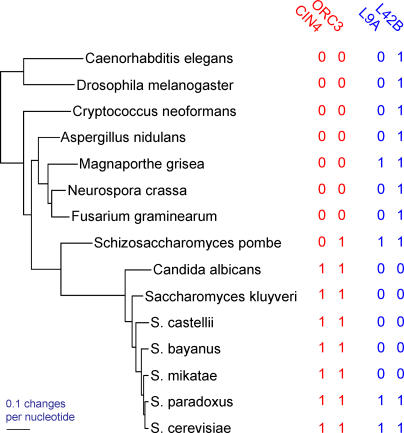
Figure 2. Phylogeny of the 15 Species Showing Two Pairs of Presence/Absence Data for Proteins in MIPS.
All nodes of the tree received 100% posterior support in an MCMC analysis (see Results). The protein pairs {CIN4, ORC3} and {L9A, L42B}, marked “1” for presence and “0” for absence, are included to illustrate probable type I (false positive) and type II (false negative) errors by the across-species method in real data (see Results, “False positives”). Probable false positive: The across-species correlation returns a significant (p = 0.0014) correlation between the pair {CIN4, ORC3}. The phylogenetic method regards this as a chance association (p = 0.13) arising from a single event of both genes being gained in the ascomycete yeasts, followed by shared inheritance (as in Figure 1). The pair {L9A, L42B} consists of two functionally linked proteins. These return a significant phylogenetic correlation (p = 0.035) owing to perhaps five correlated losses of both genes (see text). The across-species association is sensitive only to the distribution of the two proteins across the tips, and returns a non-significant result (p = 0.23). This is a probable false negative.