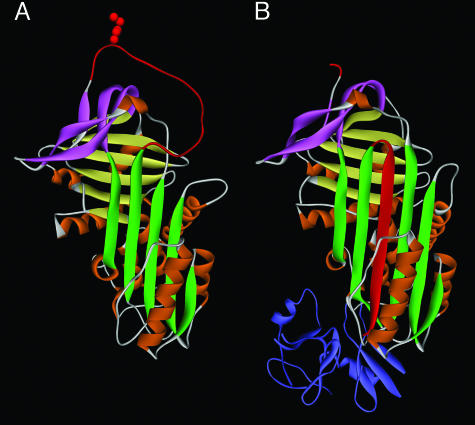
Fig. 1.
Protein structure of inhibitory serpins. (A) The native or stressed (S) conformation. The five-stranded β-sheet A is shown in green. The RCL shown at the top in red includes the specificity-determining bait “P1” residue (side chain is shown), which is exposed and free to interact with protease. In the inhibitory reaction, the protease approaches from the top, binds reversibly at the P1 site, and initiates proteolysis by forming a covalent acyl “intermediate” bond at P1 and cleaving after P1. Cleavage releases the S conformation, allowing opening of β-sheet A and rapid topological conversion to the relaxed (R) conformation. (B) The serpin–protease complex. The serpin is in the R conformation. Note that the cleaved RCL has been inserted in the center of the serpin as an additional strand in β-sheet A and has dragged the covalently bound protease (shown in blue) by >70 Å to the opposite end of the serpin. The mechanism, called “kinetic trapping,” irreversibly inactivates the protease by gross distortion of its catalytic center. Protein coordinates in A and B are from α1-antitrypsin (SERPINA1) in the native state (1QLP) and in complex with protease (1EZX), respectively. Sheet B is shown in yellow, sheet C in pink, and the nine helices in orange.