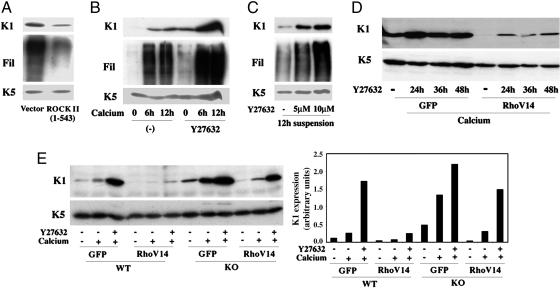
Fig. 4.
ROCK and CRIK kinases exert overlapping compensatory effects as negative regulators of differentiation. (A) Primary keratinocytes were transfected with an expression vector for a constitutively active form of ROCK II (amino acids 1–543) or empty vector control alongside an expression vector for GFP and induced to differentiate by culture in suspension for 24 h. Transfected GFP-positive cells were purified by sorting, and total cell extracts were analyzed by immunoblotting with antibodies against the indicated proteins (Fil, Filaggrin). (B) Keratinocytes were treated with the ROCK inhibitor Y27632 (5 μM) for 24 h and either kept under growing conditions or induced to differentiate by calcium for the indicated times (hours) before termination of the experiment. Total cell extracts were analyzed by immunoblotting as before. (C) Keratinocytes were treated with Y27632 at the indicated concentrations for 12 h followed by induction of differentiation by suspension culture for additional 12 h. Total cell extracts were analyzed by immunoblotting. (D) Keratinocytes were infected with recombinant adenoviruses expressing RhoV14 or GFP control for 48 h, with induction of differentiation by calcium for the last 12 h of the experiment. Cells were either untreated or treated with Y27632 (5 μM) for the indicated times (hours) before termination of the experiment. Samples were analyzed by immunoblotting with antibodies against the keratin 1 and 5 markers. (E) Keratinocytes from CRIK–/– mice (KO) and wild-type littermate controls (WT) were infected with the GFP and RhoV14 adenoviruses (multiplicity of infection, 10) for 48 h, and either kept under growing conditions or induced to differentiate by calcium for the last 12 h of the experiment. Some cultures were also concomitantly treated with Y27632 (5 μM) for the last 24 h of the experiment, as indicated. Expression of the K1 and K5 proteins was monitored by immunoblotting (Left). Data were quantified by densitometry and expressed as arbitrary units after normalization for levels of K5 expression (Right).