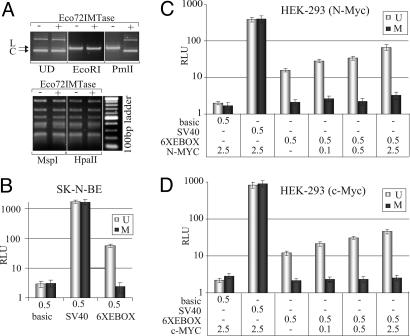
Fig. 5.
E-box methylation can affect Myc transcription activity in vivo. (A) The pGL2-6XEBOX vector, prepared from either DH10B(-) or DH10B-MT (+) E. coli strains, was separated on a 1% agarose gel and stained with ethidium bromide. Migration profiles of both plasmid forms were compared. UD, undigested; EcoRI, digestion with EcoRI used as a control; PmlI, digestion with PmlI to discriminate between methylated and unmethylated E-box sequences; MspI, digestion with MspI that cleaves the CCGG sequence regardless of cytosine methylation; HpaII, digestion with HpaII that cleaves the unmethylated CCGG sequence only. L, linear form of the plasmid; C, supercoiled form of the plasmid. (B) Luciferase activity of reporters prepared from DH10B (U, unmethylated) or DH10B-MT (M, methylated) after transfection into SK-N-BE cells. (C and D) Luciferase activity of reporters measured as a function of increased N-Myc or c-Myc expression in HEK-293 cells. The activity of the reporter firefly luciferase was normalized to that of the Renilla, used as an internal control. Relative luciferase units (RLU) were plotted on a semilogarithmic scale. Micrograms of transfected plasmids are indicated on the x axes of respective graphs.