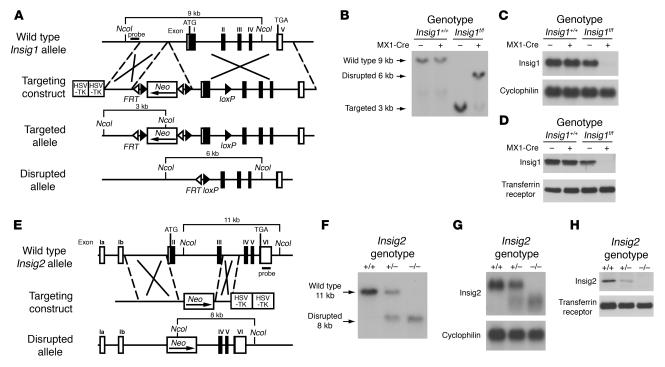
Figure 1.
Targeted disruption of Insig1 and Insig2 genes in mice. (A) Schematic of Insig1 gene-targeting strategy. Cre-mediated excision of the sequences between loxP sites deletes exon 1. The location of the probe used for Southern blot analysis is denoted by the horizontal filled rectangle labeled “probe.” (B) Representative Southern blot analysis of NcoI-digested DNA from livers of mice with the indicated genotypes that were treated with 4 intraperitoneal injections of pIpC (300 μg/injection). (C) Northern blot analysis of hepatic RNA of mice indicated in B. Total RNA from liver was pooled, and 20-μg aliquots were subjected to electrophoresis and blot hybridization with 32P-labeled cDNA probes for mouse Insig1 and mouse cyclophilin. (D) Immunoblot analysis of livers of mice indicated in B. Liver membrane fractions were prepared as described in Methods, and aliquots (45 μg) were subjected to SDS-PAGE and immunoblot analysis. (E) Schematic of Insig2 gene-targeting strategy. The Insig2 allele was disrupted by replacement of exons II and III of the Insig2 gene with a polIIsneopA expression cassette. The DNA probe used for Southern blot analysis is denoted by the horizontal filled rectangle labeled “probe.” (F) Representative Southern blot analysis of NcoI-digested tail DNA of the offspring from mating of Insig2+/− mice. (G) Northern blot analysis of hepatic RNA of mice described in F. Total RNA from livers of mice was subjected to electrophoresis and blot hybridization with 32P-labeled cDNA probes for mouse Insig2 and mouse cyclophilin. (H) Immunoblot analysis of liver membranes from mice with the indicated Insig2 genotype, as described above.