Abstract
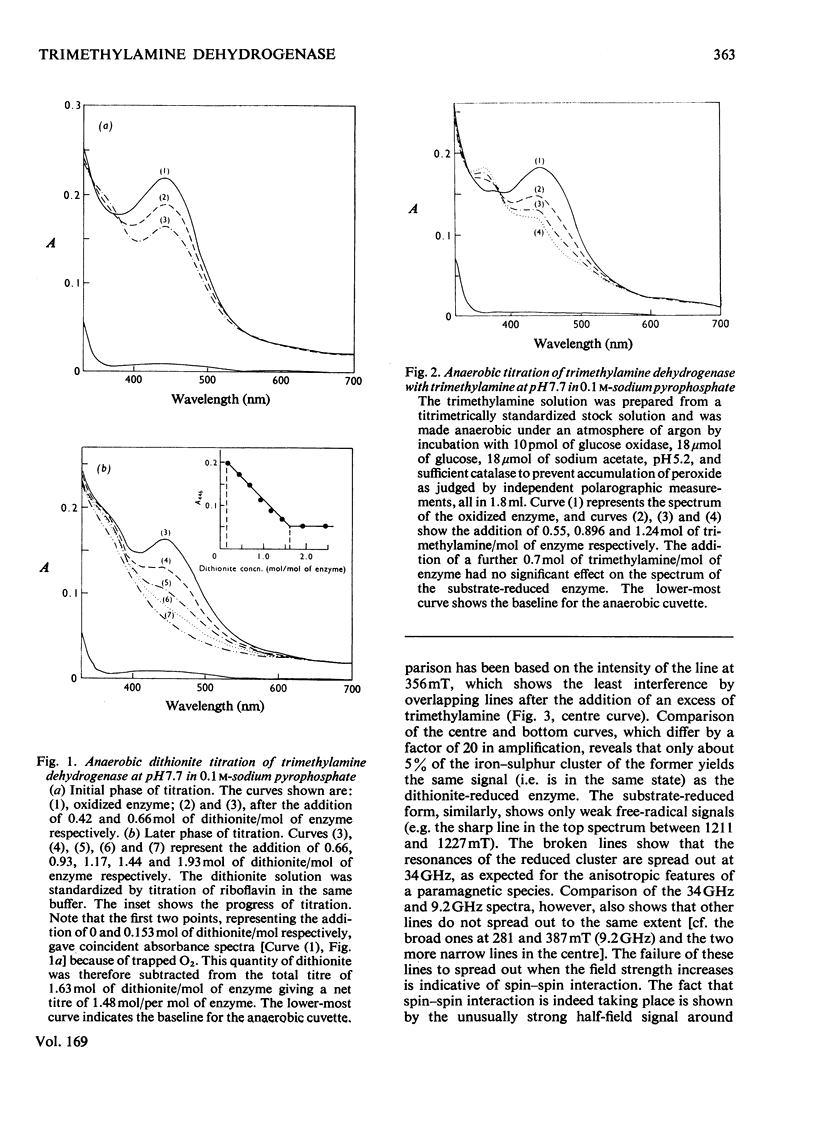
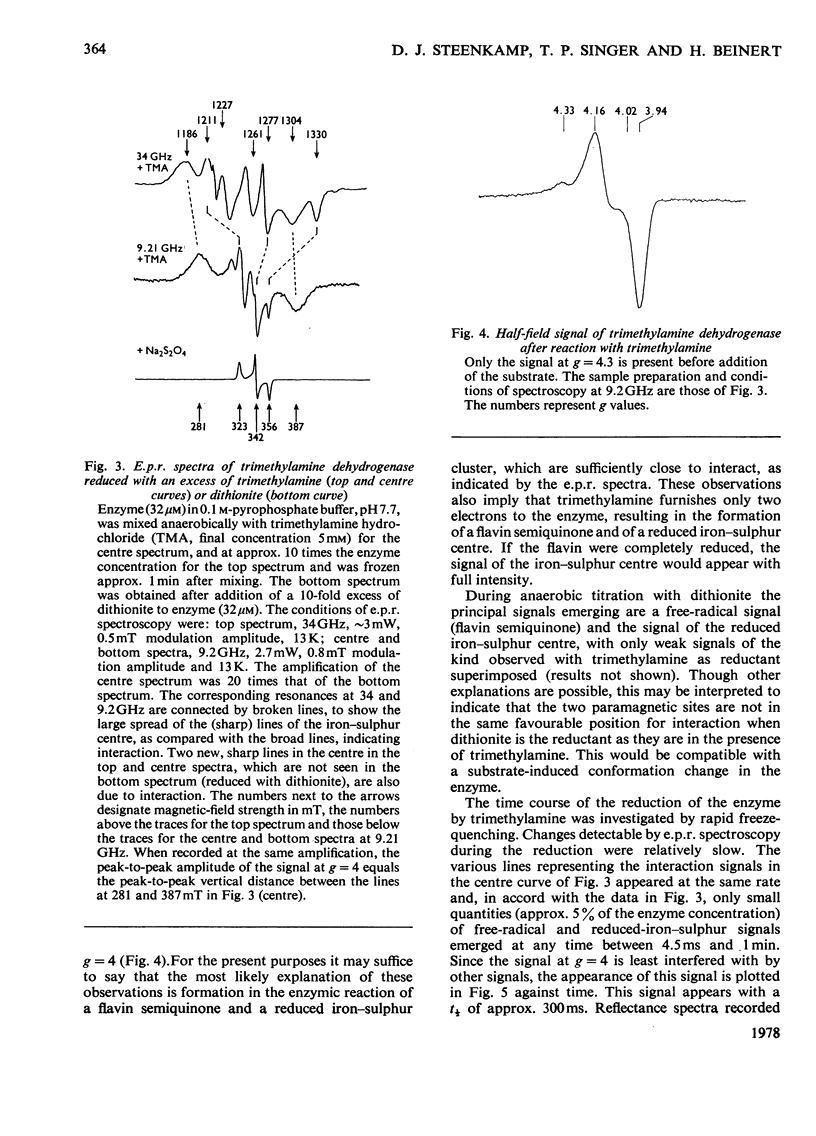
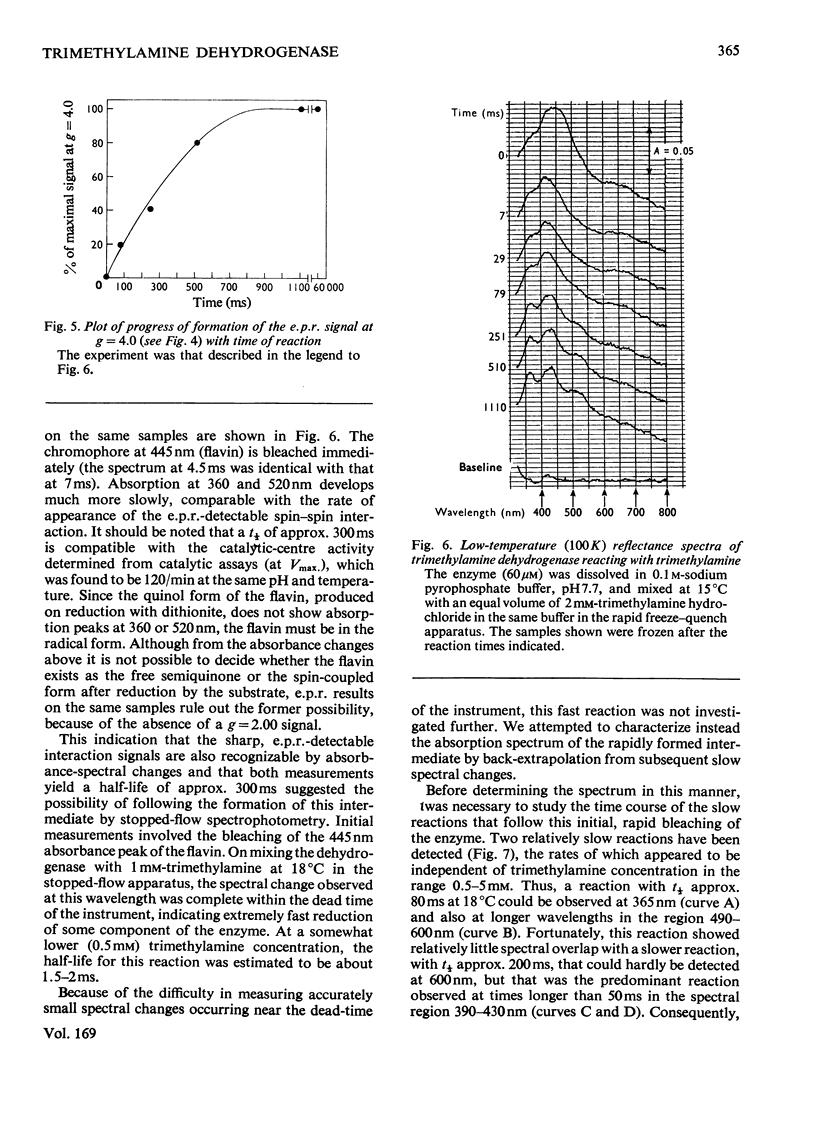
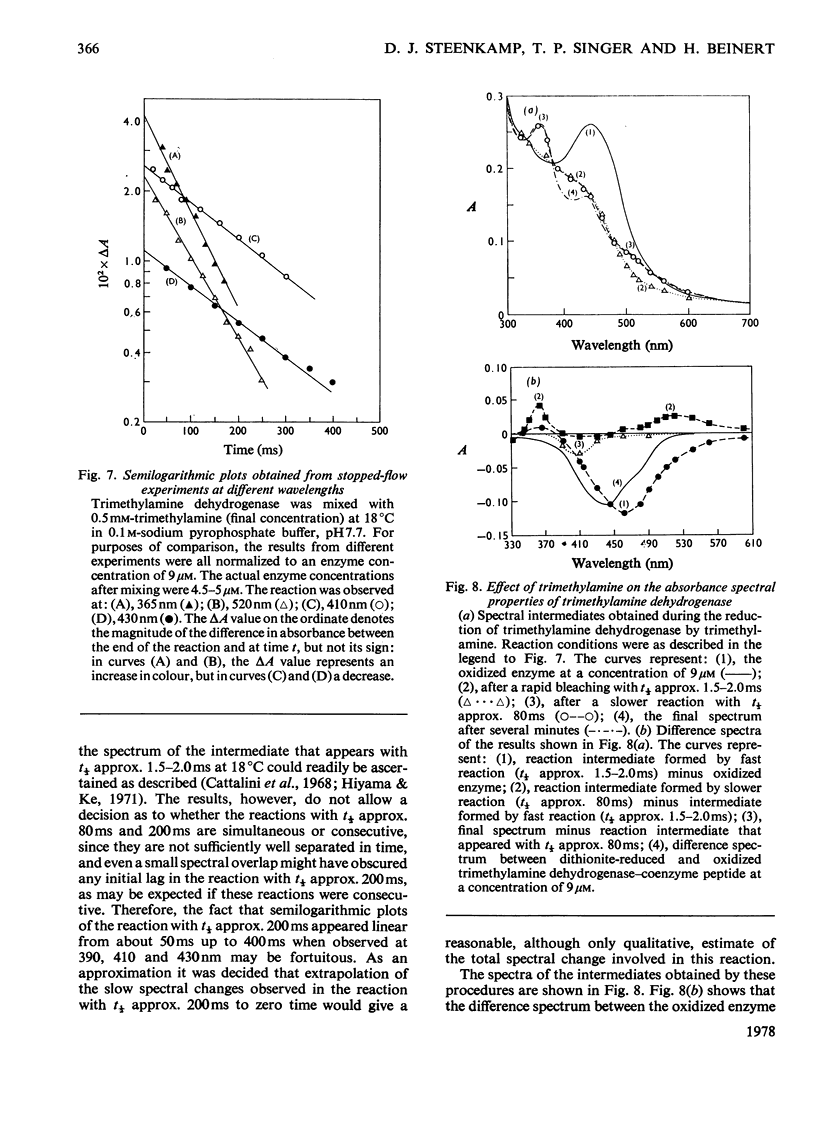
Bacterial trimethylamine dehydrogenase contains a novel type of covalently bound flavin mononucleotide and a tetrameric iron-sulphur centre. The dehydrogenase takes up 1.5mol of dithionite/mol of enzyme and is thereby converted into the flavin quinol-reduced (4Fe-4S) form, with the expected bleaching of the visible absorption band of the flavin and the emergence of signals of typical reduced ferredoxin in the electronparamagnetic-resonance spectrum. On reduction with a slight excess of substrate, however, unusual absorption and electron-paramagnetic-resonance spectra appear quite rapidly. The latter is attributed to extensive interaction between the reduced (4Fe-4S) centre and the flavin semiquinone. The species of enzyme arising during the catalytic cycle were studied by a combination of rapid-freeze e.p.r. and stopped-flow spectophotometry. The initial reduction of the flavin to the quinol form is far too rapid to be rate-limiting in catalysis, as is the reoxidation of the substrate-reduced enzyme by phenazine methosulphate. Formation of the spin-spin-interacting species from the dihydroflavin is considerably slower, however, and it may be the rate-limiting step in the catalytic cycle, since its rate of formation agrees reasonably well with the catalytic-centre activity determined in steady-state kinetic assays. In addition to the interacting form, a second form of the enzyme was noted during reduction by trimethylamine, differing in absorption spectrum, the structure of which remains to be determined.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beinert H., Ackrell B. A., Kearney E. B., Singer T. P. Iron-sulfur components of succinate dehydrogenase: stoichiometry and kinetic behavior in activated preparations. Eur J Biochem. 1975 May;54(1):185–194. doi: 10.1111/j.1432-1033.1975.tb04128.x. [DOI] [PubMed] [Google Scholar]
- Beinert H., Hansen R. E., Hartzell C. R. Kinetic studies on cytochrome c oxidase by combined epr and reflectance spectroscopy after rapid freezing. Biochim Biophys Acta. 1976 Feb 16;423(2):339–355. doi: 10.1016/0005-2728(76)90190-0. [DOI] [PubMed] [Google Scholar]
- Colby J., Zatman L. J. Purification and properties of the trimethylamine dehydrogenase of bacterium 4B6. Biochem J. 1974 Dec;143(3):555–567. doi: 10.1042/bj1430555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J., Zatman L. J. Tricarboxylic acid-cycle and related enzymes in restricted facultative methylotrophs. Biochem J. 1975 Jun;148(3):505–511. doi: 10.1042/bj1480505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D., Ballou D., Van Heuvelen A., Palmer G., Massey V. Kinetic studies on the substrate reduction of xanthine oxidase. J Biol Chem. 1973 Sep 10;248(17):6135–6144. [PubMed] [Google Scholar]
- Gillum W. O., Mortenson L. E., Chen J. S., Holm R. H. Quantitative extrusions of the Fe4S4 cores of the active sites of ferredoxins and the hydrogenase of Clostridium pasteurianum. J Am Chem Soc. 1977 Jan 19;99(2):584–595. doi: 10.1021/ja00444a044. [DOI] [PubMed] [Google Scholar]
- Hill C. L., Steenkamp D. J., Holm R. H., Singer T. P. Identification of the iron-sulfur center in trimethylamine dehydrogenase. Proc Natl Acad Sci U S A. 1977 Feb;74(2):547–551. doi: 10.1073/pnas.74.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama T., Ke B. A new photosynthetic pigment, "P430": its possible role as the priary electron acceptor of photosystem I. Proc Natl Acad Sci U S A. 1971 May;68(5):1010–1013. doi: 10.1073/pnas.68.5.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Hemmerich P., Ehrenberg A. Light absorption of flavosemiquinone metal chelates. Eur J Biochem. 1968 Jul;5(2):158–164. doi: 10.1111/j.1432-1033.1968.tb00352.x. [DOI] [PubMed] [Google Scholar]
- Olson J. S., Ballou D. P., Palmer G., Massey V. The mechanism of action of xanthine oxidase. J Biol Chem. 1974 Jul 25;249(14):4363–4382. [PubMed] [Google Scholar]
- Orme-Johnson N. R., Hansen R. E., Beinert H. Electron paramagnetic resonance-detectable electron acceptors in beef heart mitochondria. Reduced diphosphopyridine nucleotide ubiquinone reductase segment of the electron transfer system. J Biol Chem. 1974 Mar 25;249(6):1922–1927. [PubMed] [Google Scholar]
- Orme-Johnson N. R., Hansen R. E., Beinert H. Electron paramagnetic resonance-detectable electron acceptors in beef heart mitochondria. Ubihydroquinone-cytochrome c reductase segment of the electron transfer system and complex mitochondrial fragments. J Biol Chem. 1974 Mar 25;249(6):1928–1939. [PubMed] [Google Scholar]
- Steenkamp D. J., Mallinson J. Trimethylamine dehydrogenase from a methylotrophic bacterium. I. Isolation and steady-state kinetics. Biochim Biophys Acta. 1976 May 13;429(3):705–719. doi: 10.1016/0005-2744(76)90319-3. [DOI] [PubMed] [Google Scholar]
- Steenkamp D. J., Schabort J. C., Ferreira N. P. Beta-cyclopiazonate oxidocyclase from Penicillium cyclopium. 3. Preliminary studies on the mechanism of action. Biochim Biophys Acta. 1973 Jun 6;309(2):440–456. doi: 10.1016/0005-2744(73)90042-9. [DOI] [PubMed] [Google Scholar]
- Steenkamp D. J., Singer T. P. On the presence of a novel covalently bound oxidation-reduction cofactor, iron and labile sulfur in trimethylamine dehydrogenase. Biochem Biophys Res Commun. 1976 Aug 23;71(4):1289–1295. doi: 10.1016/0006-291x(76)90794-4. [DOI] [PubMed] [Google Scholar]


