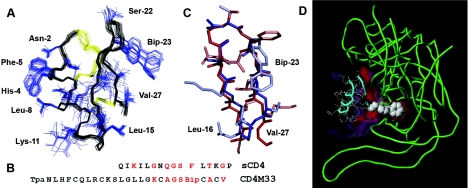
Figure 1. Three-dimensional structure of CD4M33 and its computed complex with gp120.
(A) Ensemble of 30 low energy NMR structures, superposed by using backbone atoms (in black); disulphide bridges are in yellow and side chains in blue. (B) Sequence alignment of the CDR2-like loop of CD4 and the engineered CD4M33 in the one-letter amino acid code. Critical residues are in red. (C) Structure comparison of the CD4 CDR2-loop (sequence 36–47) and the CD4M33 β-hairpin (sequence 16–27). (D) Three-dimensional model of CD4M33 (cyan) in complex with gp120 (green), computed according to the CD4–gp120 complex crystallographic structure (PDB code: 1g9m). Protruding Bip side chain engaging gp120 Phe43 cavity is represented in space-filling model and the interface in red.