Abstract
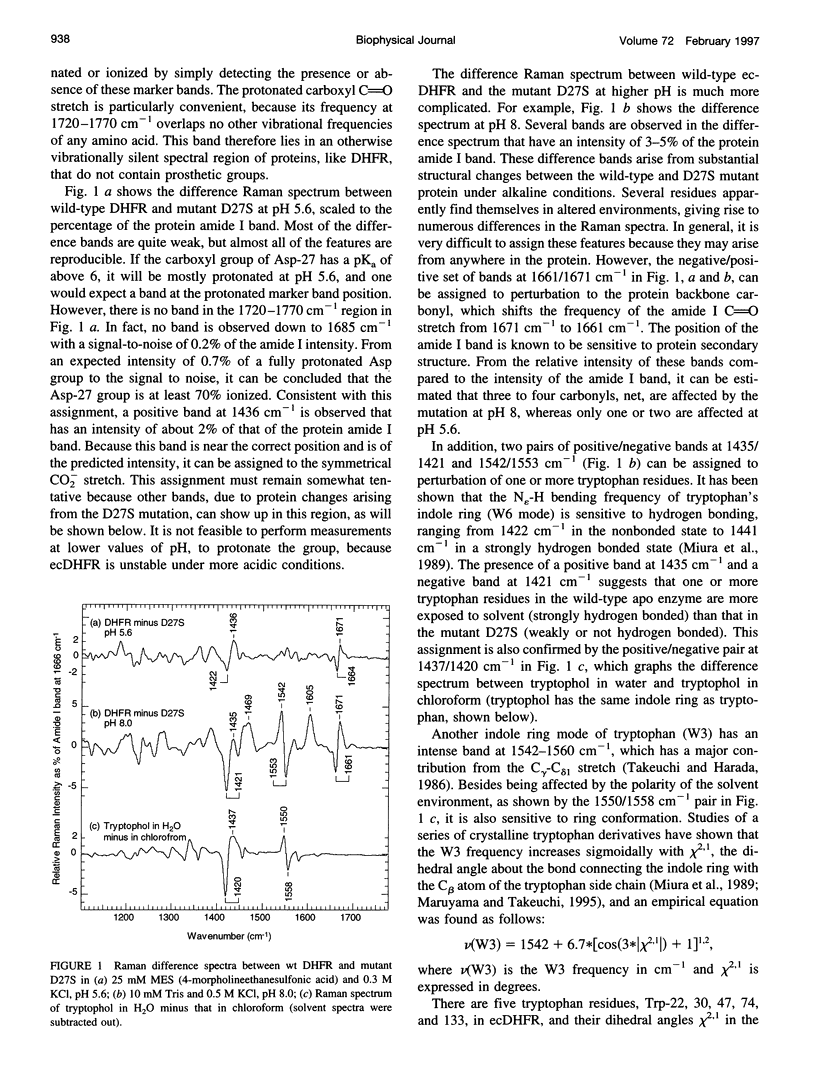
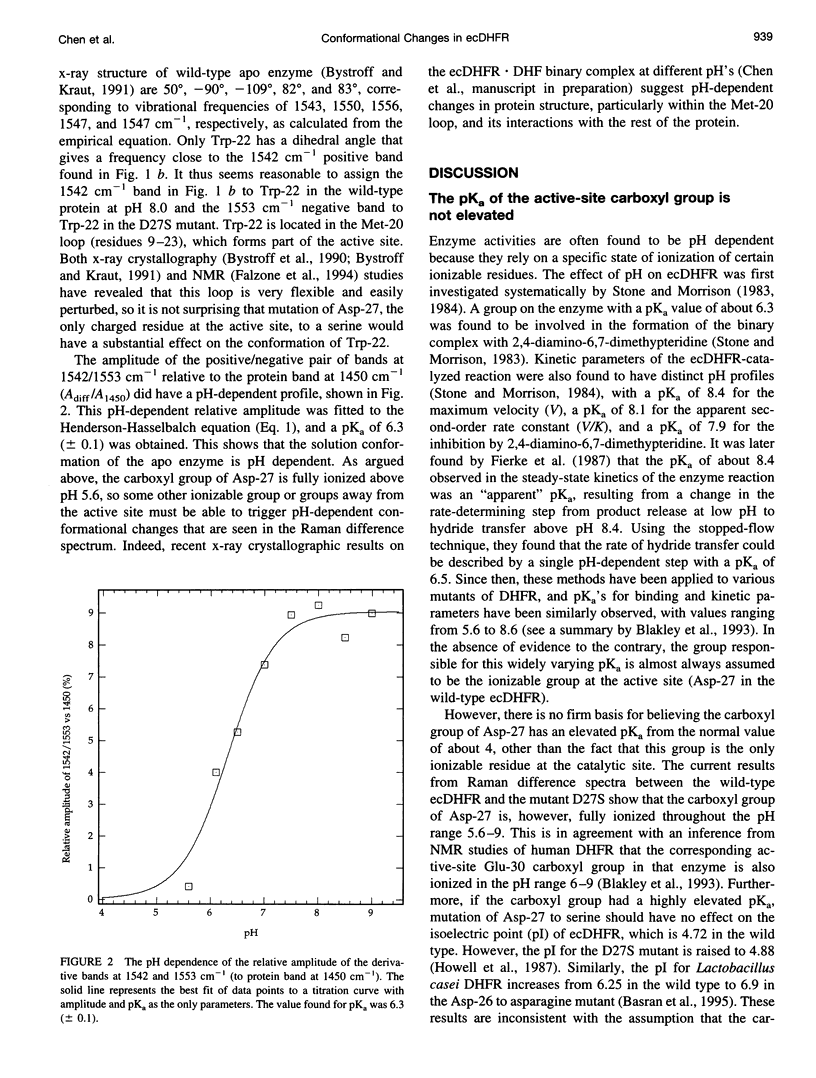
The catalytic site of all dihydrofolate reductases contains an invariant carboxylic acid, equivalent to Asp-27 in Escherichia coli dihydrofolate reductase (ecDHFR). It has been found that various kinetic and ligand binding properties of ecDHFR show a pH profile with a pKa of about 6.5. The group responsible for this pKa is often assumed to be the carboxyl group of Asp-27. To determine the ionization state of this carboxyl and its pKa, we have employed a novel method, based on Raman difference spectroscopy, to obtain its vibrational spectrum in situ. The method is general for the study of protein carboxyl groups, which are often significantly implicated in protein function and structure; this study establishes the method's limits and problems. The Raman difference spectrum between wild-type ecDHFR and the Asp-27 to serine mutant (D27S) in the pH range 5.6-9.0 has been taken. No protonation of the carboxyl group was detected, implying that its pKa is probably less than 5.0. We did, however, detect a pH dependence in the intensity of Raman bands in the difference spectrum with a pKa of 6.3, indicating that the apo enzyme undergoes a pH-dependent conformational change. Because the carboxyl group of Asp-27 at the active site is the only ionizable group in the binding site, other groups, away from the catalytic site, must be responsible for the pH behavior of ecDHFR.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J., Johnson K., Matthews R., Benkovic S. J. Effects of distal point-site mutations on the binding and catalysis of dihydrofolate reductase from Escherichia coli. Biochemistry. 1989 Aug 8;28(16):6611–6618. doi: 10.1021/bi00442a012. [DOI] [PubMed] [Google Scholar]
- Appleman J. R., Howell E. E., Kraut J., Blakley R. L. Role of aspartate 27 of dihydrofolate reductase from Escherichia coli in interconversion of active and inactive enzyme conformers and binding of NADPH. J Biol Chem. 1990 Apr 5;265(10):5579–5584. [PubMed] [Google Scholar]
- Basran J., Casarotto M. G., Barsukov I. L., Roberts G. C. Role of the active-site carboxylate in dihydrofolate reductase: kinetic and spectroscopic studies of the aspartate 26-->asparagine mutant of the Lactobacillus casei enzyme. Biochemistry. 1995 Mar 7;34(9):2872–2882. doi: 10.1021/bi00009a018. [DOI] [PubMed] [Google Scholar]
- Blakley R. L., Appleman J. R., Freisheim J. H., Jablonsky M. J. 13C and 15N nuclear magnetic resonance evidence that the active site carboxyl group of dihydrofolate reductase is not involved in the relay of a proton to substrate. Arch Biochem Biophys. 1993 Nov 1;306(2):501–509. doi: 10.1006/abbi.1993.1543. [DOI] [PubMed] [Google Scholar]
- Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. J Biol Chem. 1982 Nov 25;257(22):13650–13662. [PubMed] [Google Scholar]
- Bystroff C., Kraut J. Crystal structure of unliganded Escherichia coli dihydrofolate reductase. Ligand-induced conformational changes and cooperativity in binding. Biochemistry. 1991 Feb 26;30(8):2227–2239. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- Bystroff C., Oatley S. J., Kraut J. Crystal structures of Escherichia coli dihydrofolate reductase: the NADP+ holoenzyme and the folate.NADP+ ternary complex. Substrate binding and a model for the transition state. Biochemistry. 1990 Apr 3;29(13):3263–3277. doi: 10.1021/bi00465a018. [DOI] [PubMed] [Google Scholar]
- Callender R., Deng H. Nonresonance Raman difference spectroscopy: a general probe of protein structure, ligand binding, enzymatic catalysis, and the structures of other biomacromolecules. Annu Rev Biophys Biomol Struct. 1994;23:215–245. doi: 10.1146/annurev.bb.23.060194.001243. [DOI] [PubMed] [Google Scholar]
- Chen Y. Q., Kraut J., Blakley R. L., Callender R. Determination by Raman spectroscopy of the pKa of N5 of dihydrofolate bound to dihydrofolate reductase: mechanistic implications. Biochemistry. 1994 Jun 14;33(23):7021–7026. doi: 10.1021/bi00189a001. [DOI] [PubMed] [Google Scholar]
- Falzone C. J., Wright P. E., Benkovic S. J. Dynamics of a flexible loop in dihydrofolate reductase from Escherichia coli and its implication for catalysis. Biochemistry. 1994 Jan 18;33(2):439–442. doi: 10.1021/bi00168a007. [DOI] [PubMed] [Google Scholar]
- Fierke C. A., Johnson K. A., Benkovic S. J. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987 Jun 30;26(13):4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- Howell E. E., Villafranca J. E., Warren M. S., Oatley S. J., Kraut J. Functional role of aspartic acid-27 in dihydrofolate reductase revealed by mutagenesis. Science. 1986 Mar 7;231(4742):1123–1128. doi: 10.1126/science.3511529. [DOI] [PubMed] [Google Scholar]
- Howell E. E., Warren M. S., Booth C. L., Villafranca J. E., Kraut J. Construction of an altered proton donation mechanism in Escherichia coli dihydrofolate reductase. Biochemistry. 1987 Dec 29;26(26):8591–8598. doi: 10.1021/bi00400a015. [DOI] [PubMed] [Google Scholar]
- Li L. Y., Benkovic S. J. Impact on catalysis of secondary structural manipulation of the alpha C-helix of Escherichia coli dihydrofolate reductase. Biochemistry. 1991 Feb 12;30(6):1470–1478. doi: 10.1021/bi00220a004. [DOI] [PubMed] [Google Scholar]
- Morrison J. F., Stone S. R. Mechanism of the reaction catalyzed by dihydrofolate reductase from Escherichia coli: pH and deuterium isotope effects with NADPH as the variable substrate. Biochemistry. 1988 Jul 26;27(15):5499–5506. doi: 10.1021/bi00415a017. [DOI] [PubMed] [Google Scholar]
- Stone S. R., Morrison J. F. Catalytic mechanism of the dihydrofolate reductase reaction as determined by pH studies. Biochemistry. 1984 Jun 5;23(12):2753–2758. doi: 10.1021/bi00307a034. [DOI] [PubMed] [Google Scholar]
- Stone S. R., Morrison J. F. The pH-dependence of the binding of dihydrofolate and substrate analogues to dihydrofolate reductase from Escherichia coli. Biochim Biophys Acta. 1983 Jun 29;745(3):247–258. doi: 10.1016/0167-4838(83)90056-0. [DOI] [PubMed] [Google Scholar]
- Uchimaru T., Tsuzuki S., Tanabe K., Benkovic S. J., Furukawa K., Taira K. Computational studies on pterins and speculations on the mechanism of action of dihydrofolate reductase. Biochem Biophys Res Commun. 1989 May 30;161(1):64–68. doi: 10.1016/0006-291x(89)91560-x. [DOI] [PubMed] [Google Scholar]
- Villafranca J. E., Howell E. E., Voet D. H., Strobel M. S., Ogden R. C., Abelson J. N., Kraut J. Directed mutagenesis of dihydrofolate reductase. Science. 1983 Nov 18;222(4625):782–788. doi: 10.1126/science.6356360. [DOI] [PubMed] [Google Scholar]


