Abstract
Aims: The isolation of various genes that are expressed in a region specific manner is considered useful for research in molecular pathology. In situ hybridisation (ISH) was used in a screening procedure to isolate these genes efficiently, using colon cancer as a model.
Methods: Suppression subtractive hybridisation (SSH) between colon cancer tissue samples and corresponding non-cancerous tissues was performed. Genes showing high expression in the cancers were selected using macro-DNA array analysis. As a final screening procedure, conventional ISH was performed to isolate genes expressed specifically in colon cancers.
Results: Sixty nine clones were selected by SSH and macro-DNA array analyses. These clones were then analysed by ISH to examine their expression patterns. ISH screening revealed that all the clones screened showed more intense signals in colon cancers than in non-cancerous tissues. Among them, RACK 1, which is a protein kinase C receptor and a homologue of the G protein β subunit, was expressed intensely in colon cancer cells. RACK 1 expression was evaluated in multiple samples by ISH, and the results confirmed that RACK 1 was universally overexpressed in cells of all 11 colon cancers examined.
Conclusions: Many genes, including RACK 1, expressed in colon cancer cells can be isolated efficiently by this method, and their precise expression pattern can be evaluated. These results indicate that ISH is an excellent technique for systemic screening of genes expressed in a region specific manner.
Various human organs and tissues have specific gene expression profiles that reflect the function of each organ.1 This is also true for cancer cells, in which many genetic changes accumulate, and the elucidation of the gene expression profiles of cancer cells might help in the search for molecular mechanisms of cancer development and investigations into the biological behaviour of tumours.2,3 Attempts have been made to isolate genes overexpressed in cancer cells using methods such as differential hybridisation, differential display, and differential subtraction,4–6 all of which are based on quantitative differences in gene expression between cancerous and non-cancerous cells. With the release of the complete human genome sequence, our ability to determine and understand the specific gene expression patterns of cancer cells will increase dramatically. Conventional approaches to sporadic searching for the expression patterns of selected genes will be replaced by rapid and high-throughput technologies, using tools such as cDNA microarrays, thereby allowing a comprehensive analysis of gene expression profiles facilitating molecular mechanisms of diseases.2,3
For the study of cancer, it is important to analyse region specific gene expression, rather than analysing tissue from throughout a tumour. Understanding genes expressed in a region specific manner on a large scale may provide important clues to the mechanisms of development and disease. For example, genes expressed at the invasion front of cancer nests are likely to contribute to the process of cancer invasion, and genes expressed at either the lymphatic or venous region of cancer invasion are likely to be candidate metastasis associated genes. Two techniques that assist the region specific localisation of genes are currently available. One is the analysis of region specific specimens that are isolated by microdissection,7,8 which allows the sampling of extremely small pieces of tissue from the relevant regions, although this has some drawbacks related to sample handling and sample quantity. The other is the localisation of gene expression itself at the tissue level, using in situ hybridisation (ISH).9 ISH gives region specific signals, and permits histopathological evaluation of the entire tissue, including the region of interest, thereby allowing gene expression to be linked to the results of histopathological examination.10,11
“Understanding genes expressed in a region specific manner on a large scale may provide important clues to the mechanisms of development and disease”
We have devised a strategy using ISH for the extensive screening of genes the expression of which is increased in colon cancers. Because simple screening using random probes may be less efficient, slower, and more expensive, we combined suppression subtraction hybridisation (SSH) and cDNA macroarray analysis to enrich genes that are highly expressed in colon cancers.12,13 These genes were then used as probes for the ISH analysis.
ISH screening confirmed that higher amounts of all the clones were expressed in cancer tissue than in non-cancer tissue. In particular, we found reproducibly increased expression of the receptor for activated C kinase (RACK 1) in colon cancer.
MATERIALS AND METHODS
Tumour samples and RNA preparation
Two specimens of colon cancer (ascending colon and sigmoid colon) and two corresponding specimens of non-cancerous colonic mucosa were obtained from surgically resected materials at the National Cancer Center Hospital, Tokyo. These samples were immediately homogenised in Trizol reagent (Gibco, Grand Island, New York, USA), and total RNA was prepared from both cancer and non-cancer tissues according to the manufacturer's instructions.
For ISH, 11 specimens of colon cancer and adjacent non-cancer tissue were also obtained from materials surgically resected at the National Cancer Center Hospital. Each specimen was immediately embedded in OCT compound and snap frozen, then 5 μm thick consecutive slides were prepared.
mRNA isolation and cDNA synthesis
Samples of total RNA (1 mg) were prepared from each tissue and then mixed together. mRNA was then extracted from the pool of colon cancer total RNA using a BioMag mRNA purification kit (Polysciences Inc, Warrington, Pennsylvania, USA); the same procedure was used for non-cancer tissue. cDNA was synthesised from each mRNA sample using an anchored oligo-dT primer supplied in the polymerase chain reaction (PCR) selected cDNA subtraction kit (Clontech, Palo Alto, California, USA), according to the manufacturer's recommendations.
Generation of subtracted cDNA library using SSH
SSH was performed between colon cancer cDNA (as a tester) and the corresponding non-cancer tissue cDNA (as a driver) using the PCR select cDNA subtraction kit (Clontech), according to the manufacturer's recommendations. Then, the subtracted cDNA was ligated with the T/A cloning vector pCR II (Invitrogen, Groningen, the Netherlands) and transformed into the Escherichia coli DH5α strain in the conventional manner. Finally, the subtracted library was spread over agar plates containing ampicillin (100 μg/ml) and X-gal (50 μl of a 2 mg/ml stock solution/90 mm plate), and incubated overnight at 37°C.
Screening using reverse northern hybridisation for macro-DNA array filter
An independent recombinant colony of the subtracted library was inoculated into a sterile deep well microtitre plate with 96 wells, each containing 750 μl of LB medium and appropriate antibiotics. After overnight incubation at 37°C, each plasmid was prepared using a 96 well glass filtered plate and saturated KI solution as described previously.14 Each plasmid sample was mixed with the same volume of 10× saline sodium citrate (SSC) and blotted on to an ImmobilonTM –NY nylon membrane (Millipore, Bedford, Maryland, USA) using a dot blotter (BioRad, Hercules, California, USA). After duplicate membranes had been prepared, they were denatured in 0.5M NaOH/1.5M NaCl and then neutralised with 1.5M NaCl/0.5M Tris/HCl (pH 7.5). The subtracted colon cancer cDNAs arrayed on the membranes were prehybridised in hybridisation buffer (1mM EDTA, 0.5M NaHPO4 (pH 7.2), containing 7% sodium dodecyl sulphate (SDS), and 2.5 mg/ml salmon sperm DNA) for three hours at 65°C. The buffer was then replaced with fresh hybridisation solution containing α32P dCTP labelled cDNA probes, which were reverse transcribed from 1 μg of cancer or non-cancer tissue mRNA.15 Hybridisation was performed overnight at 65°C, and the arrays were washed under highly stringent conditions with 0.2× SSC, 0.1% SDS at 65°C. To compare the signal intensities on the autoradiography films for the duplicate membranes, the compensated differential signals between cancer tissue and non-cancer tissue cDNA fragments were detected.
ISH screening
Recombinant plasmids containing subtracted colon cancer cDNAs served as templates for ISH probe synthesis. Using universal primers at multicloning sites, we performed PCR for transcriptional units to generate antisense and sense cRNA probes. In vitro transcription was carried out with SP6 RNA polymerase (Roche Diagnostics, Mannheim, Germany) or T7 RNA polymerase (Strategene, La Jolla, California, USA) and digoxigenin labelled UTP mix (Roche Diagnostics) from these transcription unit fragment templates.
ISH was performed on 5 μm thick frozen sections.16 Briefly, slides were fixed with 4% paraformaldehyde in phosphate buffer, and then passed through 1 μg/ml proteinase K in phosphate buffer, 0.1M triethanolamine containing 1/400 (vol/vol) acetic anhydrase, and 0.2M HCl. The slides were hybridised overnight at 55°C with digoxigenin labelled cRNA probes in a mixture containing ISH buffer (Dako, Kyoto, Japan), 0.6 mg/ml yeast tRNA, and competitors against multicloning site sequences flanking the cloning fragments. Post-hybridisation washes were performed with 5× SSC, 50% formamide in 2× SSC at 55°C, 10mM Tris/HCl (pH 8.0), 0.5M NaCl-1mM EDTA containing 10 mg/ml ribonuclease A and 10 000 U/ml ribonuclease T1 at 37°C, and then 2× SSC, 0.2× SSC at 55°C. Signals were visualised with antidigoxigenin alkaline phosphatase conjugated antibody (Dako) and the chromogenic substrate nitroblue tetrazolium/5-bromo-4-chloro-3-indiol phosphate (NBT/BCIP, Roche Diagnostics).
Northern blot hybridisation
Total RNA (5 μg) from eight matched cancer and non-cancer tissue samples was electrophoresed on a 1% agarose formaldehyde gel and transferred to an Immobilon–NY nylon membrane (Millipore). Loading of equal amounts of RNA was confirmed by the staining intensity for 18S ribosomal RNA. The hybridisation conditions were the same as for reverse northern hybridisation, except that the template was 70 ng of RACK 1 cDNA fragment labelled with α32P dCTP using a High prime labelling kit (Roche Diagnostics).
DNA sequencing
Each clone that was shown to be overexpressed in colon cancer by means of ISH was sequenced from the SP6 and/or T7 promoter sequences flanking the cloning sites with the Bigdye terminator cycle sequencing ready reaction mix (Perkin Elmer, Foster City, California, USA) and analysed with an Applied Biosystems 310 genetic analyser (Perkin Elmer). Homology searches for clone identification were performed with the BLAST program at The National Center for Biotechnology Information, USA.
RESULTS
Samples and libraries
Total RNA, mRNA, and the subtracted cDNA libraries were prepared from the cancer and non-cancer mucosal specimens. The subtracted cDNA was subcloned into plasmids by T/A cloning, yielding about 10 000 independent subtracted cDNA clone libraries. The selected clones were subjected to PCR using a primer set at the multicloning site of the plasmid to amplify the insert. The length of the inserted fragment was within the range 200–1700 bp (data not shown). In parallel with this experimental system, exogenous cDNAs were added to cDNA libraries of colon cancer at a ratio of 1/10 000, and eightfold higher signals were detected in cancer cDNA probes than in non-cancer cDNA probes after SSH, suggesting that SSH yielded about an eightfold higher enrichment of cDNA (data not shown).
Isolation by SSH of genes overexpressed in colon cancers
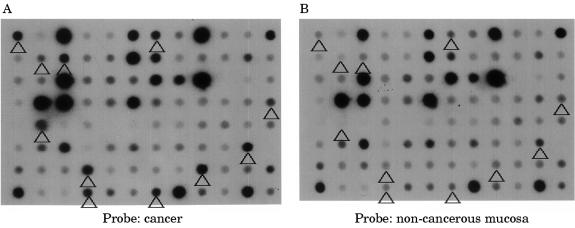
From the subtracted cDNA clone libraries, 956 clones, the expression of which was expected to be higher in colon cancer, were selected. Reverse northern hybridisation revealed that 456 (47.7%) of the clones showed stronger signals in cancer tissue cDNA probes than in non-cancer tissue cDNA probes. From these 456 clones, 69 showing a cancer/non-cancer tissue signal ratio of 5 : 1 or higher were selected (fig 1 ▶).
Figure 1.
Macro-DNA array analysis of a subtracted colon cancer cDNA library prepared by suppression subtractive hybridisation. Reverse northern hybridisation of macro arrays spotted with individual clones with α32P labelled cDNA probes synthesised from cancerous and non-cancerous tissue was performed. The spots indicated by the arrowheads show signals more than five times as intense with the cancer tissue cDNA probe (A) than with the non-cancerous mucosa cDNA probe (B).
ISH screening of genes overexpressed in colon cancers
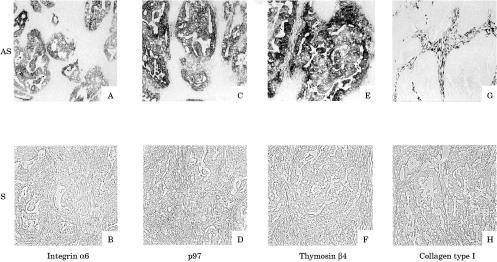
All 69 clones selected by SSH and the subsequent macro-DNA array were screened by ISH. Figure 2 ▶ shows representative clones identified by sequencing. Among these clones, integrin α6, P97, and thymosin β4 provided signals that were detected in cancer cells only.
Figure 2.
In situ hybridisation screening of colon cancers. The representative clones are shown, and the names identified by sequencing are indicated below. The signals were detected in cancer cells for selected clones such as integrin α6 (A), p97 (C), and thymosin β4 (E) with each antisense probe. In contrast, signals were detected in interstitial cells, but not in cancer cells, for collagen type I (G) with the antisense probe. Hybridisation signals were absent on consecutive sections treated with sense probes (B, D, F, and H). Original magnification, ×10. AS, antisense; S, sense.
In the case of collagen type I, on the other hand, signals were detected in the cancer stroma, but not in cancer cells. Thus, although there was variation in the localisation of expression in the cancer region, all clones showed stronger signals in cancer regions than in non-cancer regions.
Identification of genes overexpressed in colon cancers
All 69 clones were sequenced and compared with a database of known sequences using the BLAST program. All clones were matched to known genes including several expressed sequence tags (table 1 ▶). These included a highly frequent repetition (50 of 69) of various non-specific crossreacting antigens (NCAs). Although mitochondrial DNA and NADH dehydrogenase subunit 2 were detected in three of 69 clones and two of 69 clones, respectively, all of the other clones were unique.
Table 1.
Summary of cDNA clones overexpressed in colon cancers selected by suppression subtractive hybridisation and macro-DNA array
| Identity | Frequency |
| Non-specific crossreacting antigen | 50 |
| Mitochodrial DNA | 3 |
| NADH dehydrogenase subunit 2 | 2 |
| Ribosomal protein S 19 | 1 |
| Ribosomal protein S 4 | 1 |
| Ribosomal protein L 27 | 1 |
| H3 histone, family 3A | 1 |
| JTV-1 | 1 |
| Guanidine nucleotide binding protein (G protein), bate polypeptide 2-like 1 (RACK 1) | 1 |
| CD24 antigen | 1 |
| Thymosin, β4 | 1 |
| Integrin α6 | 1 |
| Prothymosin, α | 1 |
| Collagen, type 1, α 2 | 1 |
| p97 (eukaryotic initiation factor 4G) | 1 |
| map 3p22, 9.65 cR from CHLC.GATA 87B02 repeat region | 1 |
| cDNA FLJ 20676 fis, clone KAIA 4294 | 1 |
Analysis of RACK 1 as a gene overexpressed in colon cancers
To confirm the validity of our screening system, the expression of RACK 1—an interesting model molecule detected in our study—was examined further in 11 colorectal cancer specimens.
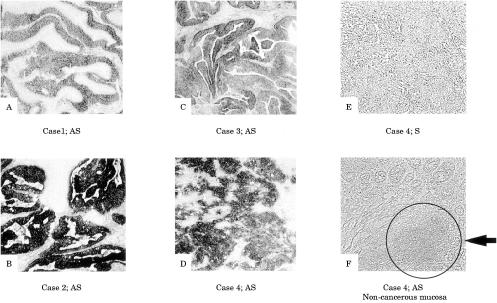
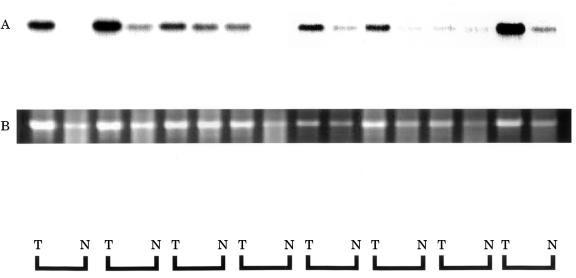
Expression of RACK 1 was detected by ISH in all 11 cases of colorectal cancer, as shown in fig 3 ▶, and its expression was strong in cancer cells. In non-cancer mucosal specimens, signals were detected in lymphoid follicles, but not in epithelial cells. The results of northern hybridisation also confirmed in multiple samples that the expression of RACK 1 mRNA was stronger in cancer regions than in non-cancer regions (fig 4 ▶). Quantitative real time reverse transcription PCR (RT-PCR) also demonstrated about twofold stronger expression of RACK 1 in cancer regions than in non-cancer regions (data not shown).
Figure 3.
RACK 1 as an overexpressed molecule in colon cancers. In situ hybridisation was performed on 11 specimens of colon cancer. RACK 1 expression was detected in the cancer cells of all 11 colon cancers. Some representative tissues are shown (A, B, C, and D). A hybridisation signal was absent on the consecutive section treated with a sense probe (E). Apart from the signal from the cancer cells, we found a signal in a lymphoid follicle in the non-cancer mucosa, as indicated by the arrow (F). Original magnification, ×10.
Figure 4.
Northern hybridisation autoradiography for RACK 1 expression. Equivalent amounts of total RNA (5 μg) were loaded and blotted. RACK 1 mRNA expression was higher in cancer tissue (T) than in non-cancer tissue (N) in all cases (A). Ethidium bromide staining of 18S ribosomal RNA is shown as the control for loading of the same quantity (B).
DISCUSSION
We have developed a strategy for isolating genes expressed in a region specific manner through ISH screening. Simple screening using random probes may be less efficient, slower, and more expensive. Therefore, we attempted to increase the efficiency of this approach by using a combination of SSH, which has been reported to enrich genes,12,13 and macro-DNA array. The advantage of enrichment is apparent from the fact that about half of the clones selected by SSH exhibited increased gene expression in the cancer region by macro-DNA array analysis. In our present study, screening by ISH of all selected clones detected stronger signals in cancer than in non-cancer regions, indicating that the combination of SSH and macro-DNA array was useful. However, because we adopted the criterion of a fivefold or higher difference between signals obtained using the cancer cDNA probe and non-cancer cDNA probe to assess the results obtained by the macro-DNA array, clones that were overexpressed in the cancer overall were preferentially selected. Thus, genes expressed at the invasion front were likely to be missed. Because SSH enriches gene expression, as is evident from the results of reverse northern hybridisation, it is expected that genes expressed in a region specific manner would be obtained by random ISH screening using as many probes as possible prepared directly from the clones obtained by SSH. In fact, large scale random ISH screening has begun to be used in developmental biology to investigate time and region specific gene expression.9 Large scale random ISH is also considered to offer great promise in the investigation of region specific gene expression for clarifying the pathogenesis of various human diseases, including cancer.
“Large scale random ISH screening has begun to be used in developmental biology to investigate time and region specific gene expression”
In our study, the length of subtracted cDNA in clones obtained by SSH that showed cancer specific expression was within the approximate range 200–1700 bp. Although the theoretical length of this fragment was 256 (44) bp, which was thought to be the average size after cleavage with the four base recognition site restriction enzyme, Rsa I, many longer fragments were obtained. This may have been the result of the acquisition of longer fragments with higher priority than shorter fragments.6 In general, fragmentation is used for preparing probes for ISH when gene fragments are long.17 However, in our present study, because the fragments approximated the theoretical size of 256 bp, we prepared probes directly from subtracted cDNA, bypassing the fragmentation process. Although several fragments greater than the theoretical size were included, all these fragments, even without fragmentation, functioned adequately as probes for ISH. This indicates that screening by ISH using SSH derived subtracted cDNA clones as probes is a satisfactory and practical method.
In 1999, Hufton et al reported the isolation of differentially expressed genes in primary colorectal cancers by SSH of cancers and non-cancer specimens and the subsequent use of a DNA macroarray.13 NCA, which is a major member of the carcinoembryonic antigen related gene family, was commonly detected in their study and in ours. The detection of NCA was repeated in four of 45 samples in their study, whereas the rate of repetition was much higher (50 of 69) in our study. This difference in the repetition frequency seems to be attributable to differences in the specimens, the efficiency of gene enrichment by SSH, and the criterion of clone selection in the macro-DNA array. The fact that the NCA clone alone was obtained repeatedly may indicate sample variation in colorectal cancers, and the need to use a larger number of specimens to correlate the expression of many genes to pathology and the degree of malignancy.
In our present study, we mounted each frozen section on a single glass slide, and carried out hybridisation on each preparation with a single sense or antisense probe for ISH. We adopted this method, rather than using the formalin fixed and paraffin wax embedded preparations commonly used in histopathology, in an attempt to obtain stronger signals.18 Although clear signals were obtained, much time and labour were required from the preparation of frozen sections to the execution of ISH. To realise high throughput detection, it would be desirable to establish a multisample ISH screening method that allows parallel processing of numerous samples, and basic studies with this aim are currently under way in our laboratory. Methods for fixing and storing samples, and for handling paraffin wax embedded samples, are issues that must be investigated to deal effectively with large numbers of samples.
ISH screening of multiple samples in our present study resulted in the isolation of RACK 1, reported to be a G protein β subunit.19 It has been shown that RACK 1 is involved in protein–protein interactions and binds particularly to integrins β1, β2, and β5 and the SH2 domain of Src.20,21 It has also been reported that when RACK 1 binds to the SH2 domain of Src, the tyrosine kinase activity is selectively inhibited.21 Although RACK 1 has many bioactivities, as described above, the expression of this molecule in human colon cancers remains unknown. Recently, RACK 1 was isolated by differential display between bovine cord forming and monolayer forming endothelial cells, and was shown to be upregulated in angiogenesis, being expressed in large vessels, but not cancer cells, in colon cancer.22 However, in our study, RACK 1 was found to be universally overexpressed in colon cancer cells.
Using ISH for screening, RACK 1 signals in the cancer region were confined to cancer cells, and in the non-cancer mucosa, signals were detected only in lymphoid follicles and not in epithelial cells. This also explains why RACK 1 was selected as a subtracted clone, because strong expression was present throughout the region of cancer cells. The expression of RACK 1 in lymphocytes has been reported previously,23 and is compatible with its expression in the lymphoid follicles of non-cancer mucosa in our present study. To determine the expression of the protein, we performed immunohistochemical staining for RACK 1 on 11 colon cancers and corresponding non-cancer mucosae. Immunoreactivity for RACK 1 was detected in the cytoplasm of cancer cells and also in lymphoid follicles in non-cancer mucosae in similar amounts to that of the mRNA (data not shown). Stronger expression of RACK 1 mRNA in cancer than in non-cancerous regions was confirmed by both northern hybridisation and quantitative real time RT-PCR, indicating that the ISH signal reflects the degree of expression. Use of ISH for screening is advantageous in that it allows comprehensive evaluation, including correlation with the histopathological picture and region specific gene expression, as compared with northern hybridisation and quantitative real time RT-PCR, which provide only quantitative information.
Take home messages.
We have developed a strategy for isolating genes expressed in a region specific manner through in situ hybridisation (ISH) screening
Many genes, including RACK 1, expressed in colon cancer cells can be isolated efficiently by this method, and their precise expression pattern can be evaluated
It is expected that ISH will be widely applicable to functional genomics in the coming post genome era
In summary, ISH screening is a very useful procedure that enables relatively easy isolation of genes expressed in a region specific manner and the analysis of region specific gene expression. ISH is therefore expected to be widely applicable to functional genomics in the coming post genome era. In the current situation, where there is an increasing need to link information about expression at the molecular level to clinicopathological information, this screening method may prove to be a valuable tool.
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Health and Welfare and for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government. This study was also supported by the Programme for Promotion of Fundamental Studies in Health Science of the Organisation for Pharmaceutical Safety and Research of Japan. AS is the recipient of Research Resident fellowship from the Foundation for Promotion of Cancer Research in Japan.
Abbreviations
ISH, in situ hybridisation
NCA, non-specific crossreacting antigen
PCR, polymerase chain reaction
RT, reverse transcription
SDS, sodium dodecyl sulphate
SSC, saline sodium citrate
SSH, suppression subtractive hybridisation
REFERENCES
- 1.Watson JD. The human genome project: past, present and future. Science 1990;248:44–9. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 3.Clark EA, Golub TR, Lander ES, et al. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000;406:532–5. [DOI] [PubMed] [Google Scholar]
- 4.Elvin P, Kerr IB, McArdle CS, et al. Isolation and preliminary characterization of cDNA clones representing mRNAs associated with tumour progression and metastasis in colorectal cancer. Br J Cancer 1988;57:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan RJ, Ford HL, Fu Y, et al. Drg-1 as a differentiation-related, putative metastatic suppression gene in human colon cancer. Cancer Res 2000;60:749–55. [PubMed] [Google Scholar]
- 6.Diatchenko L, Lau YC, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A 1996;93:6025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fend F, Quintanilla-Martinez L, Kumar S, et al. Composite low grade B-cell lymphomas with two immunophenotypically distinct cell populations are true biclonal lymphomas. Am J Pathol 1999;154:1857–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito A, Kanai Y, Maesawa C, et al. Disruption of E-cadherin-mediated cell adhesion systems in gastric cancers in young patients. Jpn J Cancer Res 1999;90:993–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komiya T, Tanigawa Y, Hirohashi S. A large-scale in situ hybridization system using an equalized cDNA library. Anal Biochem 1997;254:23–30. [DOI] [PubMed] [Google Scholar]
- 10.Komiya T, Tanigawa Y, Hirohashi S. Cloning of the gene gob-4, which is expressed in intestinal goblet cells in mice. Biochim Biophys Acta 1999;1444:434–8. [DOI] [PubMed] [Google Scholar]
- 11.Komiya T, Tanigawa Y, Hirohashi S. Cloning and identification of the gene gob-5, which is expressed in intestinal goblet cells in mice. Biochem Biophys Res Commun 1999;255:347–51. [DOI] [PubMed] [Google Scholar]
- 12.Yang GP, Ross DT, Kuang WW, et al. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res 1999;27:1517–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hufton SE, Moerkerk PT, Brandwijk R, et al. A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett 1999;463:77–82. [DOI] [PubMed] [Google Scholar]
- 14.Fujii G, Tsuchiya R, Ezoe E, et al. Analysis of nuclear localization signals using a green fluorescent protein-fusion protein library. Exp Cell Res 1999;251:299–306. [DOI] [PubMed] [Google Scholar]
- 15.Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A 1984;81:1991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oka H, Kameya T, Sato Y, et al. Significance of growth hormone-releasing hormone receptor mRNA in non-neoplastic pituitary and pituitary adenomas: a study by RT-PCR and in situ hybridization. J Neurooncol 1999;41:197–204. [DOI] [PubMed] [Google Scholar]
- 17.Cox KH, Deleon DV, Angerer LM, et al. Detection of mRNA in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol 1984;101:485–502. [DOI] [PubMed] [Google Scholar]
- 18.Braton AJL, Pearson RCA, Najlerahim A, et al. Pre- and postmortem influences on brain RNA. J Neurochem 1993;61:1–11. [DOI] [PubMed] [Google Scholar]
- 19.Ron D, Chen CH, Caldwell J, et al. Cloning of an intracellular receptor for protein kinase C: a homologue of the β subunit of G proteins. Proc Natl Acad Sci U S A 1994;91:839–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liliental J, Chang DD. Rack 1, a receptor for activated protein kinase C, interacts with integrin β subunit. J Biol Chem 1998;273:2379–83. [DOI] [PubMed] [Google Scholar]
- 21.Chang BY, Conroy KB, Machleder EM, et al. RACK 1, a receptor for activated C kinase and a homolog of the β subunit of G proteins, inhibits activity of Src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol 1998;18:3245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berns H, Humar R, Hengeret B, et al. RACK1 is up-regulated in angiogenesis and human carcinomas. FASEB J 2000;14:2549–58. [DOI] [PubMed] [Google Scholar]
- 23.Geijsen N, Spaargaren M, Raaijmakers JAM, et al. Association of RACK 1 and PKCβ with the common β-chain of the IL-5/IL-3/GM-CSF receptor. Oncogene 1999;18:5126–30. [DOI] [PubMed] [Google Scholar]