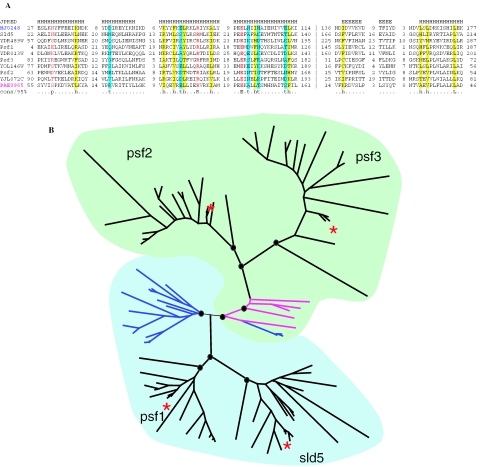
Figure 4.
Evolution of the GINS family. (A) Multiple alignment of the selected GINS proteins. Sequences are denoted by gene names: Sld5, Psf1, Psf3, Psf2—experimentally characterized GINS proteins from Xenopus laevis (70); YDR489W, YDR013W, YOL146W, YJL072C—orthologous proteins from S.cerevisiae (71); MJ0248—homolog from the euryarchaeon Methanocaldococcus jannaschii; PAE0965—homolog from the crenarchaeon Pyrobaculum aerophilum. The positions of the first and the last residue of the aligned region in the corresponding protein are indicated for each sequence. The numbers within the alignment represent poorly conserved inserts that are not shown. The vertical dashed line separates the permuted region. The colouring is based on the consensus (calculated for all sequences in the alignment) shown underneath the alignment; h/yellow indicates hydrophobic residues (ACFILMVWYHRK), t/cyan indicates turn-forming residues (ASTDNVGPERK), p/red indicates charged residues (STEDKRNQH), positions with identical amino acids are boldfaced. The secondary structure was predicted using the JPRED program (81). H indicates α-helix, E indicates extended conformation (β-strand). (B) Schematic representation of the phylogenetic tree of the GINS family. The representation is based on a maximum likelihood tree of 97 sequences of GINS family reconstructed using ProtML program. Nodes with bootstrap support >70% are marked by circles. Euryarchaeal branches are shown in blue, and the Crenarchaeal branches are shown in magenta. The two coloured areas denote the two permuted forms of the protein. Branches corresponding to the Sld5, Psf1, Psf3, Psf2 proteins from X.laevis are marked by red asterisks.