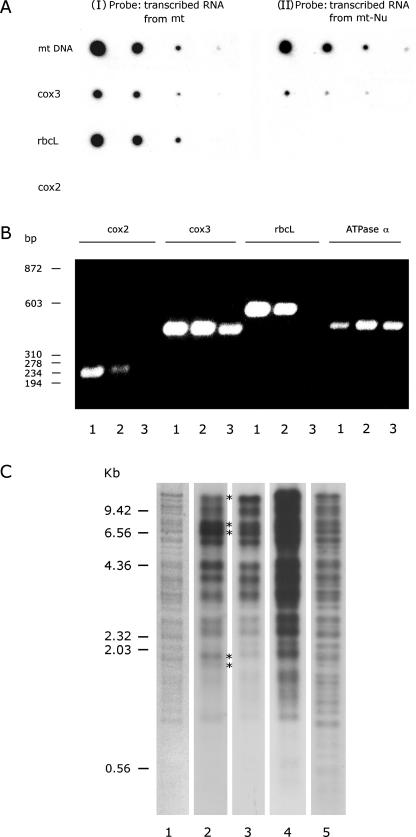
Figure 10.
Characterization of mitochondrial-nucleoid RNA synthesis in vitro. (A) The specificity of newly synthesized RNA. (I): As a control, a newly synthesized, 32P-labeled RNA isolated from 1 mg protein equivalent of purified mitochondria was used to probe a duplicate membrane described in (II). (II): A nucleoids sample isolated from 1 mg protein equivalent of mitochondria was subjected to RNA synthesis in vitro in the presence of [α-32P]UTP. The newly synthesized, 32P-labeled RNA was then isolated to probe serially diluted DNA templates immobilized on a membrane. The DNA templates are: mtDNA and PCR-amplified gene fragment of cox 3, rbc L, and cox 2. (B) Specificity of mitochondrial-nucleoid RNA. The RNA templates used for reserve PCR were derived, from whole mung bean seedlings (lane 1), mitochondria (lane 2) and isolated mitochondrial nucleoids (lane 3). Four sets of primers (see Materials and Methods) each designed to amplify a coding region of nucleus-encoded cox 2, mitochondrion-encoded cox 3, chloroplast-encoded rbc L and mitochondrion-encoded α-subunit of ATPase were used for standard reverse PCRs. The product of these 12 individual PCRs was analyzed on an agarose gel. The size of double-stranded DNA markers is indicated on the left. Approximately a billion-fold amplification revealed the presence of contaminating chloroplast RNA transcripts in the mitochondrial RNA samples (lane 2 in rbcL sector) isolated from mitochondria. (C) A comparison of newly synthesized RNA and DNA products of mitochondrial nucleoids. Two nucleoids samples, each isolated from 5 mg protein equivalent of mitochondria, were subjected to RNA synthesis in the presence of [α-32P]UTP and to DNA synthesis in the presence of [α-32P]dCTP. The newly synthesized RNA was then purified and used to probe a DNA blot containing EcoRI-fragments of CsCl-gradient-purified mtDNA (lane 2). An identical blot was probed by the newly synthesized, 32P-labeled DNA (lane 3). Asterisks shown in lane 2 indicate the nonstoichiometrical bands resulting from hybridization of newly synthesized RNA with mtDNA-EcoRI fragments. Lane 4 (48 h exposure) came from the same blot as lane 3 (5 h exposure). EcoRI fragments of 32P-labeled, newly synthesized DNA isolated from mitochondria are shown in lane 5 as a control. The EtBr-stained, mtDNA-EcoRI-fragment banding pattern is shown in lane 1.