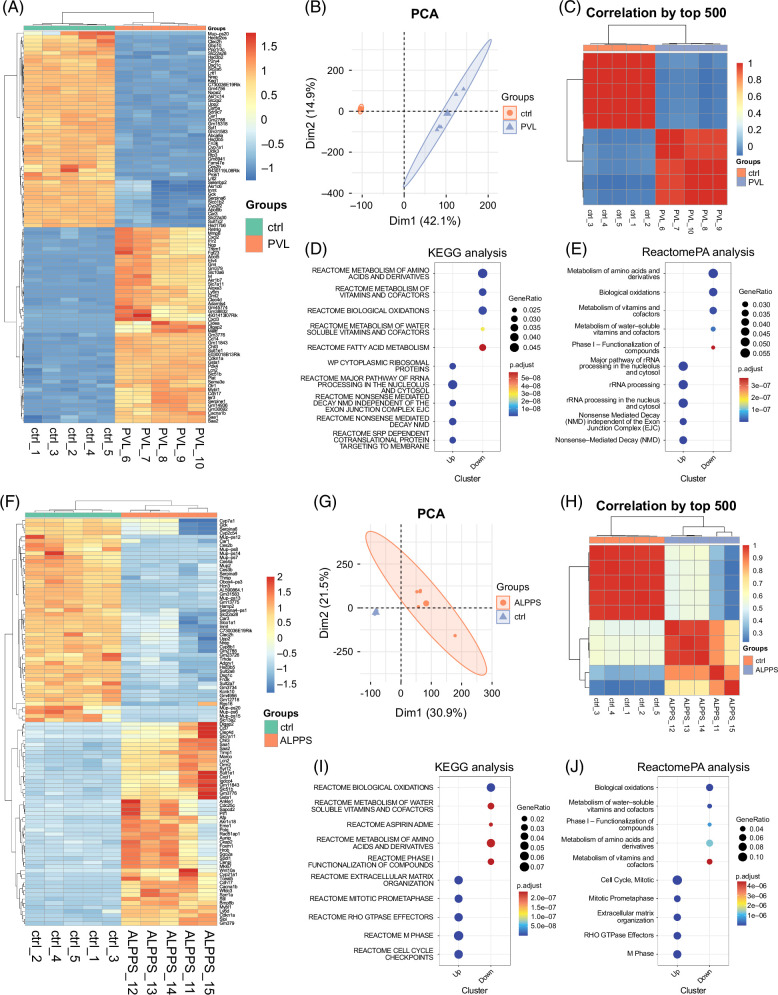
FIGURE 2.
Characteristics of liver regeneration between ALPPS and PVL based on RNA-seq analysis. (A) Heatmap shows the differentially expressed mRNAs (DEGs) between the ctrl (sham) and ALPPS model. (B) PCA based on RNA-seq data. Different colors indicate the different groups. (C) Correlation analysis of the 6 RNA-seq samples. The color represents the correlation coefficient between each sample. (D) KEGG enrichment analysis for DEGs (ALPPS versus ctrl). (E) Reactome pathway enrichment analysis for DEGs (ALPPS versus ctrl). (F) Heatmap shows the differentially expressed mRNAs (DEGs) between the ctrl (sham) and ALPPS model. (G) PCA based on RNA-seq data. Different colors indicate the different groups. (H) Correlation analysis of the six RNA-seq samples. The color represents the correlation coefficient between each sample. (I) KEGG enrichment analysis for DEGs (ALPPS versus ctrl). (J) Reactome pathway enrichment analysis for DEGs (ALPPS versus ctrl). Abbreviations: ALPPS, associating liver partition and portal vein ligation for staged hepatectomy; DEG, differentially expressed gene; PCA, principal component analysis.