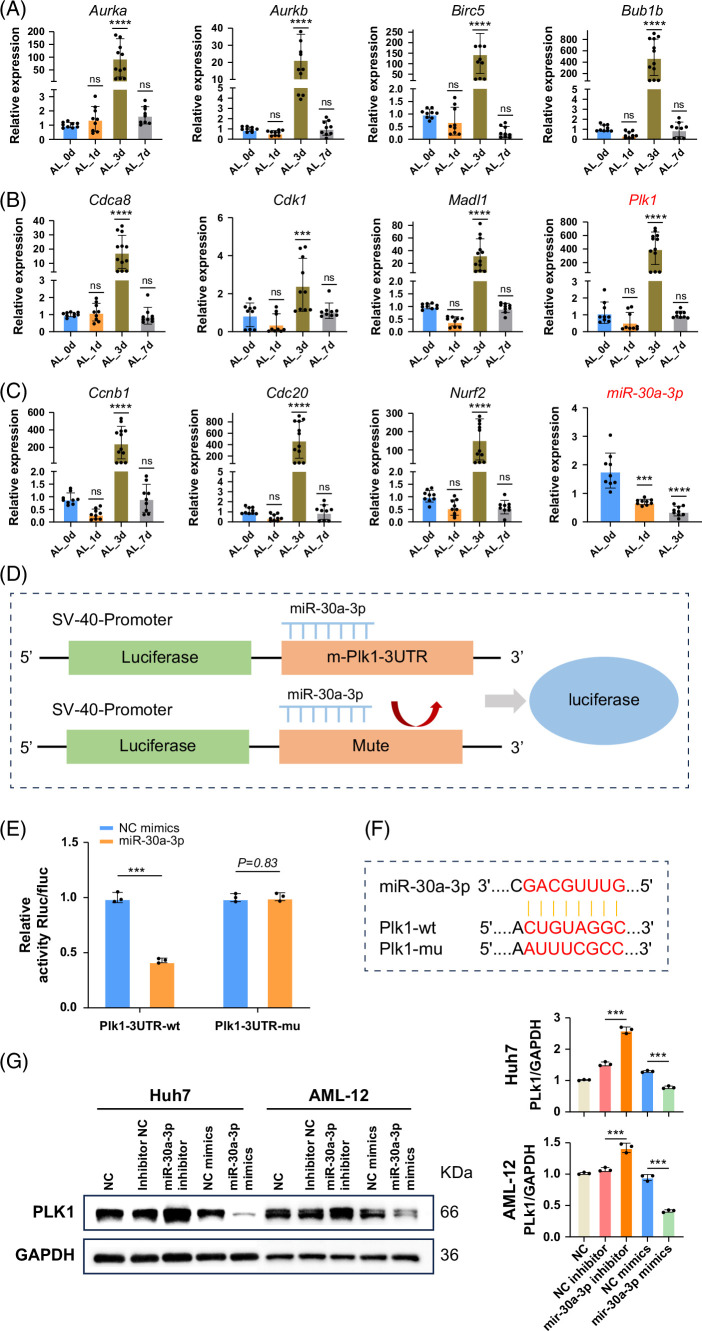
FIGURE 7.
RT-PCR and dual-luciferase assays validated the interaction between Plk1 and miR-30a-3p. (A–C) We selected 11 in 30 gene databases that significantly altered regeneration-related genes for RT-PCR validation and observed significant upregulation of previously reported hepatocyte proliferation-related genes such as Cdk1 and Ccnb1 on day 3 after ALPPS. Similarly, Plk1 exhibited the same trend. In addition, a downregulation trend of miR-30a-3p was observed at this time point. Data are shown as means ± SEM. ***p < 0.001, ns = not statistically significant, compared to the Sham group, n = 9. (D) Schematic diagram of the dual-luciferase assay illustrating the binding interaction between miR-30a-3p and Plk1. (E) The dual-luciferase assay demonstrated the binding interaction between Plk1 and miR-30a-3p, which can be reversed by mutating the Plk1 binding site. Data are shown as means ± SEM. ***p < 0.001. NC mimics versus miR-30a-3p group, n = 3. (F) Predicted binding sites of miR-30a-3p for both wild-type and mutant Plk1. (G) The expression of Plk1 was evaluated in both the overexpression and inhibition systems. In vitro transfection of the Huh7 and AML-12 cell lines was performed to simulate the microRNA overexpression and inhibition systems. Western blot results demonstrated that the inhibition of microRNA significantly enhanced the expression of Plk1, whereas the overexpression of microRNA significantly reduced Plk1 expression. Data are shown as means ± SEM. ***p < 0.001. NC mimics versus miR-30a-3p mimics, inhibitor NC versus miR-30a-3p inhibitor group, n = 3. Abbreviations: ALPPS, associating liver partition and portal vein ligation for staged hepatectomy; NC, negative control; Plk1, polo-like kinase 1.