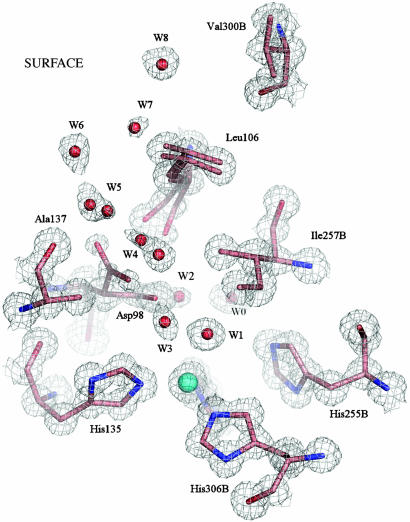
Fig. 1.
The T2Cu site of the resting state enzyme at 0.9 Å resolution is shown with 2Fobs – Fcalc electron density contoured at 1.2σ. The site is well ordered, with isotropic B factors of 5.3 Å2 for the Cu2+ atom and an average of 5.4 Å2 for the NE2 atoms of the three ligating histidine ligands (100, 135, 306B), coordinated at 1.99 Å. A water molecule, W1, is the fourth Cu ligand, at 2.2 Å with a B factor of 5.9 Å2. The Ile257B residue from the second monomer provides a steric constraint (W1 to Ile257B is ≈3.4 Å) on the substrate binding site (8). A well ordered network of water molecules, W3–W8, leads from the substrate-binding site to the protein surface along the probable route of nitrite entry. W2, W4, and W5 have partial or dual occupancy, depending on the orientation of the Asp-98 side chain, either toward the substrate-binding pocket (proximal orientation) or into the substrate-entry channel (gatekeeper orientation). In the resting enzyme, the gatekeeper orientation has lower occupancy, as suggested by the weaker electron density for this position of the Asp-98 side chain. The Leu-106 residue also exhibits a dual conformation, corresponding to the different positions of Asp-98, W4 and W5. Water molecule W0 belongs to the proton-transfer water network.