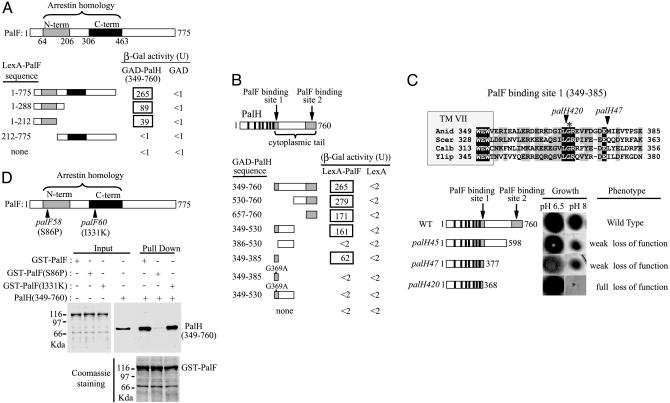
Fig. 3.
Mutational analysis of the interaction between PalF and PalH. (A and B) Two-hybrid mapping of the PalF- and PalH-interacting domains. (A) LexA-PalF fusions containing indicated PalF residues were tested for interaction with GAD-PalH (349-760) or GAD alone. N-term (shaded bar) and C-term (solid bar) indicate PalF PFAM arrestin N-terminal and C-terminal domains, respectively. (B) GAD-PalH fusions containing indicated PalH residues were tested for interaction with LexA-PalF or LexA alone. Solid bars indicate the positions of PalH-predicted transmembrane helix (8), and shaded bars indicate the two PalF-binding sites. (C) Phenotypic analysis of palH mutations affecting the PalF-binding sites. The sequence alignment including A. nidulans PalH and S. cerevisiae, Y. lipolytica, and C. albicans Rim21 shows the PalF-binding site located immediately downstream of the seventh PalH TM helix (TM VII). (Upper) Fully conserved (black shading) or similar (BLOSUM62, gray shading) residues are shown. Arrowheads indicate positions of the truncation sites in palH420 and palH47 mutant proteins. Fully conserved PalH Gly-369, crucial for PalF-PalH interaction, is denoted by an asterisk. Numbers at the ends indicate residue positions. (Lower Right) A phenotypic analysis of palH mutant strains (8, 18) grown on synthetic media at the indicated pH values. (Lower Left) Schematic representation of truncated proteins. Filled boxes and solid bars are as in B. Numbers on the right show the final in-frame encoded residues of the truncated proteins. (D) In vitro binding of palF58 (S86P) and palF60 (I331K) proteins to the C-terminal region of PalH. Pull-down assays were as in Fig. 1.