Abstract
Deletion of the 234-bp core element of the DNase I hypersensitive site 3 (5′HS3) of the locus control region (LCR) in the context of a human beta-globin locus yeast artificial chromosome (β-YAC) results in profound effects on globin gene expression in transgenic mice. In contrast, deletion of a 2.3-kb 5′HS3 region, which includes the 234-bp core sequence, has a much milder phenotype. Here we report the effects of these deletions on chromatin structure in the beta-globin locus of adult erythroblasts. The 234-bp 5′HS3 deletion abolished histone acetylation throughout the β-globin locus; recruitment of RNA polymerase II (pol II) to the LCR and beta-globin gene promoter was reduced to a basal level; and formation of all the 5′ DNase I hypersensitive sites of the LCR was disrupted. The 2.3-kb 5′HS3 deletion mildly reduced the level of histone acetylation but did not change the profile across the whole locus; the 5′ DNase I hypersensitive sites of the LCR were formed, but to a lesser extent; and recruitment of pol II was reduced, but only marginally. These data support the hypothesis that the LCR forms a specific chromatin structure and acts as a single entity. Based on these results we elaborate on a model of LCR chromatin architecture which accommodates the distinct phenotypes of the 5′HS3 and HS3 core deletions.
The human β-globin gene cluster lies in chromosome 11 and spans ∼100 kb. The locus contains a distal enhancer located at the 5′ end of the cluster, termed the locus control region (LCR), and five functional genes that are arranged according to their expression during ontogeny (51). Although transcription of the globin genes can be detected by various sensitive assays in the absence of the LCR, the LCR is indispensable for determining the physiological level of expression of the globin genes in vivo (1, 18). In addition to transcriptional enhancer activity, the LCR is able to confer upon the globin genes position-independent, copy number-dependent expression at ectopic chromatin sites, suggesting that it possesses chromatin opening activity (18). In transgenic mice the LCR enhances globin gene expression by 100- to 1,000-fold (18). Deletion of the mouse LCR by homologous recombination reduces expression of the β-globin genes to basal levels (1), confirming the importance of LCR enhancer activity in situ. Deletion of the LCR does not result in closure of the locus chromatin (1, 11, 46), as would have been anticipated based upon transgenic studies (18). Thus, in addition to the LCR, another cis regulatory element(s) may be involved in opening the β-globin locus chromatin domain.
The LCR contains multiple DNase I hypersensitive (HS) sites (15, 18, 58). Classic enhancer activity of 5′ HS site 2 (5′HS2) can be detected by transient transfection assays (39, 59), whereas enhancer activity of 5′HSs 3 and 4 can only be measured by stable transfection assays or with transgenic mice (17, 44, 47, 53), suggesting that the enhancer function of 5′HS3 and 4 is associated with chromatin modification. Using enhancer blocking and boundary assays, 5′HS5 shows chromatin insulating activity in vivo (33, 34, 55, 61, 63). However, deletion by homologous recombination of the LCR sequences including 5′HS5 does not have a considerable effect on globin gene expression in the mouse (3, 12), suggesting that its in situ function remains unknown. 5′HS1 has no enhancer activity, and no phenotype was observed in adult individuals who carried a 5′HS1 deletion mutation (27). Each 5′HS site contains a core sequence spanning approximately 200 to 300 bp; these core sequences are highly conserved during evolution (reviewed in references 19, 32, and 35). The distances between the cores are also roughly conserved, separated by 3 to 4 kb (35). The cores are characterized by multiple binding motifs for erythroid and non-erythroid transcription factors and co-activators, including Ap1/NF-E2, GATA1, and Sp1/EKLF (reviewed in reference 51).
While deletion of the entire LCR dramatically disrupts globin gene expression (1), the removal of individual 5′HS sites and their flanking sequences (approximately 2 to 3 kb) by homologous recombination results in an only 20 to 30% decrease of expression, suggesting that the effect of each 5′HS on globin gene expression is additive, rather than synergistic (3, 4, 13, 21). This conclusion is also supported by studies of transgenic mice. Mutations that remove a 2- to 3-kb sequence of 5′HS2 or 5′HS3 individually in the context of a human β-globin locus yeast artificial chromosome (β-YAC) construct have only a moderate effect on expression of the human globin genes in transgenic mice (36, 41, 42). In contrast, deletions that remove 200 to 300 bp of the 5′HS2 or 3 or 4 core sequences individually produce catastrophic effects on globin gene expression (6, 7, 37, 38). For example, deletion of the 234-bp 5′HS3 core abolished ɛ-globin gene expression during embryonic erythropoiesis but did not affect γ-globin gene expression in this developmental stage (38). However, this mutation eliminated γ-globin gene expression during fetal definitive erythropoiesis. Expression of the β-globin gene was also considerably decreased, and it was sensitive to integration position (38).
These observations raise the question as to how the removal of a large segment of a 5′HS produces a mild phenotype whereas the removal of only a 5′HS core produces dramatic alteration of globin gene expression. The concept of the LCR “holocomplex” was postulated to account for these seemingly discrepant data (6, 7, 62). The holocomplex hypothesis suggests that the LCR is a single structural entity, with the cores forming an active site and the core flanking regions maintaining the conformation. Removal of a large segment of a 5′HS site does not significantly interfere with function; the remaining 5′HSs can adopt an alternate, albeit slightly less functional conformation and active site. Thus, the impact on gene expression would be moderate. However, when only a 5′HS core is missing, the original holocomplex structure is maintained, but the active site is severely disturbed resulting in a dominant negative mutation. The LCR cannot adopt a new conformation, because the intact core-flanking sequences maintain the conformation of the wild holocomplex. Thus, the effect on globin gene expression is catastrophic. These profoundly different phenotypes reveal intrinsic properties of the LCR structure and its function. To delineate this specific property of the LCR, we analyzed histone acetylation, 5′HS formation, and recruitment of trans-acting factors in wild-type, 2.3-kb 5′HS3 deletion, and 5′HS3 core deletion β-YAC transgenic mice adult erythroblasts. Our study showed that the 234-bp 5′HS3 deletion impairs formation of HSs, abolishes histone acetylation, and inhibits RNA polymerase (pol II) recruitment at the globin locus. The 2.3-kb HS deletion also globally affects chromatin structure and recruitment of trans-acting factors but to a much lesser extent. These phenotypes can be accounted for by a modified LCR holocomplex hypothesis.
MATERIALS AND METHODS
Materials.
The production of transgenic mice carrying the wild-type, 2.3-kb HS3, or 234-bp HS3 deletion βYAC construct was described previously (38, 42). Rabbit polyclonal antibodies against histone H3 acetylated at lysines 9 and 14 (catalog no. 06-599) and histone H4 acetylated at lysines 5, 8, 12, and 16 (06-866) were purchased from Upstate Biotechnology (Lake Placid, NY); rabbit polyclonal antibodies against pol II (N-20, sc-899) and NF-E2 (C-19, sc-291) and goat polyclonal antibody against GATA-1 (C-20, sc-1233) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). DNase I was purchased from Worthington Biochemical (Lakewood, NJ). LightCycler FastStart DNA Master SYBR Green I PCR kit was from Roche Diagnostics (Indianapolis, IN), and QuantiTect SYBR Green PCR kit was from QIAGEN (Valencia, CA). PCR was performed using a LightCycler (Roche) or a DNA Engine Opticon 2 (MJ Research, Waltham, MA).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed using phenylhydrazine-treated spleens from adult transgenic mice. Histone acetylation and recruitment of polII, GATA1, and NF-E2 were measured by a real-time PCR-based ChIP assay (9), with the following modifications. Single-cell spleen suspensions were prepared from 10- to 12-week-old βYAC transgenic mice 4 days after onset of phenylhydrazine-induced hemolytic anemia. The mouse autoimmune regulator (mAire) gene, which is minimally expressed in adult mice splenic cells, and the murine βmaj-globin gene were selected as internal control. In vivo regulation of these two endogenous mouse genes presumably is not affected by expression or integration of transgenes; thus, ratios of histone acetylation or binding of trans-acting factors in these two genes was assumed to be constant between different transgenic samples. Data sets in which was the mAire/βmaj ratio within 0.16 ± 0.05 were considered to be good-quality experiments and were included in result analyses. Immunoprecipitations (IP) were performed at least three times on different days. Three dilutions of each DNA were utilized, and the PCR readings of the three dilutions had to be within the range of the standard DNA curve. The PCR data were used only when the PCR efficiency of the standard DNA in the experiment was 1.9 ± 0.1. All data were expressed as the ratio of the PCR readings of a given primer set over the internal control, and standard deviation was calculated.
DNase I hypersensitivity assay.
Single-cell spleen suspensions were prepared from phenylhydrazine-treated 10- to 12-week-old β-YAC transgenic mice. Isolation of nuclei, DNase I digestion, and Southern blot hybridization were performed as described previously (9). The restriction enzymes employed for genomic DNA digestion and the probes used for Southern blot hybridization are listed (see Fig. 2).
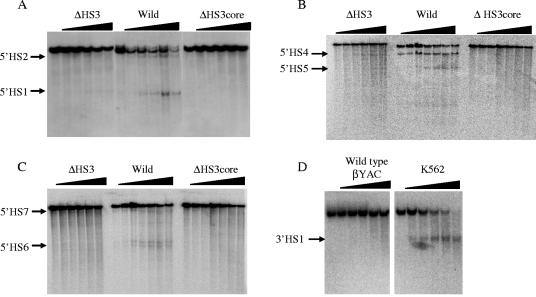
FIG. 2.
Effects of the HS3 mutants on formation of DNase I hypersensitive sties. The βYAC constructs are labeled on the upper part on each panel. (A) Detection of HSs 1 and 2. DNA was digested with restriction enzyme NdeI; the blot was hybridized with a 3′ probe. The parental band is 8.7 kb; subbands are 2.1 kb (HS1) and 6.9 kb (HS2). (B) Detection of HSs 4 and 5. DNA was digested with restriction enzyme ScaI; the blot was hybridized with a 5′ probe. The parental band is 11.5 kb; subbands are 4 kb (HS5) and 6.4 kb (HS4). (C) Detection of HSs 6 and 7. DNA was digested with restriction enzyme ScaI; the blot was hybridized with a 3′ probe. The parental band is 12 kb; subbands are 2.4 and 2.5 kb (HS6) and 9 kb (HS7). (D) Detection of 3′HS1. DNA was digested with restriction enzyme ScaI; the blot was hybridized with a 3′ probe. The parental band is 9 kb; the subband is 2.5 kb.
Primers.
PCR primer sequences are available on request.
RESULTS
We have previously reported two types of βYAC transgenic mice carrying a 5′HS3 mutation. One carries a 234-bp deletion of the 5′HS3 core (38); the other carries a 2.3-kb deletion of the entire 5′HS3 region (42) encompassing the 5′HS3 core sequence and flanking sequences highly conserved between human, mouse, goat, and rabbit (reviewed in reference 19). The phenotype of transgenic mice carrying the large 5′HS3 deletion βYAC was relatively mild; expression of the ɛ-globin gene was reduced to 70% of that seen with wild-type β-YAC animals in the yolk sac, levels of γ-globin gene expression were close to that of the wild type in the yolk sac and fetal liver, and β-globin gene expression in fetal and adult erythroid cells was slightly decreased. The average level of β-globin gene expression from all transgenic lines was approximately 70% of the wild-type control level in day 14 fetal liver and in adult blood (42). In contrast, deletion of the 234-bp 5′HS3 core sequence resulted in catastrophic effects on globin gene expression. Expression of the ɛ-globin gene was completed abolished in the yolk sac, while there was no effect on γ-globin gene expression. In the fetal liver γ-globin gene expression was abolished and β-globin gene expression was significantly reduced. Position-sensitive and reduced expression of the β-globin gene was observed in adult erythroid cells. The average level of β-globin gene expression from all transgenic lines was approximately 30% of the wild-type control level in adult blood (ranging from 3% to 59%) (38). In this study, we used a 5′HS3 core deletion transgenic line in which the β gene was expressed at 3%, which was the lowest level among the all established lines in the original study.
Profiles of histone acetylation.
It is thought that gene expression is regulated in part by histone acetylation. We determined the distributions of histone acetylation across the entire β-globin locus chromatin domain in adult erythroblasts of wild-type and mutant 5′HS3 βYAC transgenic mice. Histone acetylation was measured by a real-time PCR-based ChIP assay (9). We found that while a profile of histone acetylation could be measured by ChIP assay, it was an arduous task to make the ChIP data comparable between different samples: although a high level of precision of PCR readings was achieved, the exponential nature of PCR compromised the accuracy of the measurements. To control the quality of ChIP assays, we introduced the following modifications. We used the ratio of histone acetylation levels of the mouse mAire over βmaj-globin gene promoters as an indicator of ChIP assay quality. The acetylation levels of these two endogenous mouse genes presumably are not affected by transgenes. Thus, acceptable data sets from different experiments were required to have a consistent mAire/βmaj ratio. Data from each set of primers were included only when the PCR amplification efficiency of the standard DNA was within the range of 1.9 ± 0.1. At least three independent immunoprecipitations were performed, and these were done on different days. Each immunoprecipitated DNA was quantitated by real-time PCR at three different dilutions. By conducting all these quality controls, we were able to make semi-quantitative comparisons between the wild-type and mutant 5′HS3 β-globin loci.
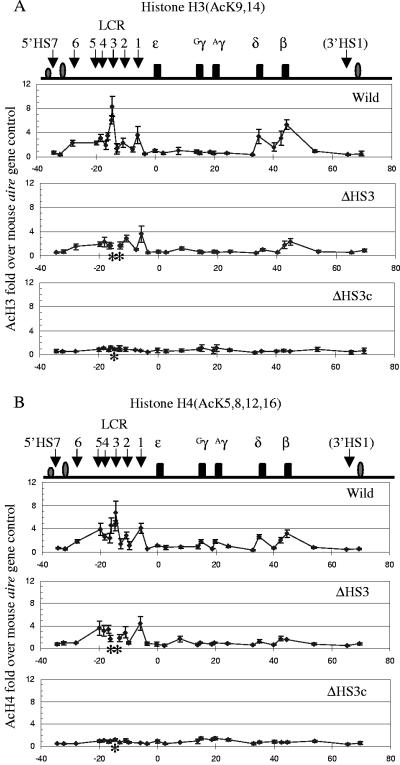
Figure 1 shows the profiles of histone H3 (panel A) and histone H4 (panel B) acetylation over an approximately 100-kb region of the β-globin locus in wild-type, Δ5′HS3, and Δ5′HS3 core transgenic mice. In each panel the top box depicts the distribution of histone acetylation in the wild-type βYAC, the middle box shows the Δ5′HS3 βYAC data, and the bottom box displays the Δ5′HS3 core βYAC results. The general acetylation profiles for histones H3 and H4 are very similar for the same construct (compare panels A and B). For example, the wild-type βYAC mice were characterized by two heavily acetylated regions encompassing the LCR and the δ/β-globin gene regions. The distribution of histone acetylation in the LCR was characterized by the peaks of histone acetylation that co-localized with the core sequences of 5′HS1, -2, -3, and -4. Histone acetylation was reduced to a lower level in the regions between the 5′HS cores. These results suggest that the trans-acting factors bound at the 5′HS cores are likely responsible for the recruitment of histone acetyltransferases. The δ- and β-globin gene promoters, which are expressed during the adult stage of development, were highly acetylated, while the region between the two genes was lightly acetylated. The approximately 40 kb of chromatin between 5′HS1 and the δ-globin gene, encompassing the silenced ɛ- and γ-globin genes, was not acetylated or was lightly acetylated. Unlike the 5′HS sites in of the LCR, 5′HS7 and 3′HS1 were not acetylated, suggesting that the two sites have properties distinct from the LCR 5′HS sites.
FIG. 1.
Profiles of histone H3 and H4 acetylation in the human β-globin locus. (A) The distribution of K9- and -14-acetylated histone H3 in adult erythroblasts of transgenic mice carrying wild-type (upper box), 2.3-kb HS3 deletion (middle box), or 234-bp HS3 core deletion (lower box) βYAC construct. The top line diagram shows the human β-globin locus with positions of the genes and HSs indicated (the drawing is to scale). The numbers labeled on the x axis are sequence coordinates, with zero set at the canonical cap site of the ɛ gene. The asterisks on the x axis indicate the deleted sequences in the mutant constructs. The y axis shows severalfold increases of acetylated histone H3 at the indicated positions compared with the mouse aire gene promoter results. Standard variation is shown as small vertical bars at each measurement point. (B) The distribution of K5-, -8-, -12-, and -16-acetylated histone H4 in the β-globin locus. See the legend of panel A for details.
The middle boxes in the two panels of Fig. 1 show histone H3 and H4 acetylation profiles for the Δ5′HS3 βYAC transgenic mice. Similar profiles were observed for histone H3 and H4 acetylation in the mutant locus. Compared to the wild-type control, the 2.3-kb 5′HS3 deletion did not considerably alter the profile, except for a mild decrease of acetylation level in the LCR and the δ- and β-globin gene regions. These changes correlate with the mild decrease of β gene expression in the Δ5′HS3 βYAC mice. In the wild-type βYAC mice the highest level of histone acetylation in the LCR was co-localized with the 5′HS3 core. This peak disappeared in the Δ5′HS3 βYAC mice due to the absence of the sequence.
The bottom boxes in Fig. 1A and B show histone acetylation profiles for the Δ5′HS3 core βYAC mice. The 234-bp 5′HS3 core deletion resulted in catastrophic effects on histone acetylation in the β-globin locus; the acetylation level of the entire region was reduced to the basal level. The impact of this deletion was not limited to the region surrounding 5′HS3, but the deletion abolished histone acetylation at the every 5′HS core in the LCR. This effect extended through the δ- and β-globin gene region that is located 40 to 50 kb downstream to the mutation.
As revealed when these data are taken together, removal of the 2.3-kb 5′HS3 mildly reduced the level of histone acetylation across the locus but did not change the general acetylation profile of the β-globin locus. In contrast, the 234-bp 5′HS3 core deletion abolished histone acetylation across the entire locus.
Formation of LCR DNase I hypersensitive sites.
The LCR is characterized by the presence of multiple DNase I hypersensitive sites (15, 18, 58). To study the impact of the 5′HS3 deletion mutations on chromatin structure, we estimated HS formation in the LCR region. Determination of 5′HS1 and -2 results by Southern blot hybridization is shown in Fig. 2A, and that of 5′HS4 and -5 is shown in Fig. 2B. The results demonstrated that the 5′HS sites in the LCR were formed in wild-type βYAC mice (the middle sections of Fig. 2A and B). In the 2.3-kb Δ5′HS3 βYAC mice these four HS sites were detectable (the left sections of Fig. 2A and B), but the intensity of the bands was fainter than those seen with the wild-type βYAC transgenic mice, suggesting that the absence of the 2.3-kb 5′HS3 sequence impaired formation of other HS sites in the LCR. Deletion of the 234-bp 5′HS3 core severely impaired (5′HS2 and 5′HS4) or abolished (5′HS1 and 5′HS5) formation of the HS sites (right sections of Fig. 2A and B). Although Southern blot hybridization is not a quantitative assay, the distinct differences in intensity of the bands between the three constructs suggests that the negative effect of the 234-bp 5′HS3 core deletion on other HS formation is much more severe than that of the 2.3-kb 5′HS3 deletion.
The effect of the 5′HS3 deletions on HS formation was not confined to the LCR. Formation of 5′HS6, which is located −28 kb 5′ to the ɛ-globin gene, was partially inhibited in the 2.3-kb 5′HS3 deletion βYAC mice; and this site was not formed in the 234-bp 5′HS3 core deletion βYAC mice (Fig. 2 C). However, formation of 5′HS7 was only slightly affected by either the larger or the core HS3 deletion, suggesting that spread of the effect of the 5′HS3 deletions on HS formation is constrained somewhere between 5′HS6 and 5′HS7. 3′HS1 was not formed in wild-type βYAC mice (Fig. 2D, right panel). As a positive control, formation of the 3′HS1 in K562 cells is shown in Fig. 2D, left panel. The 3′HS1 sequence was intact in the βYAC construct (data not shown). The lack of 3′HS1 in the wild-type βYAC mice impeded assessment of the extent of the effects of the 5′HS3 mutants in the 3′ portion of the locus. The absence of 3′HS1 formation was also reported in mini-β-locus transgenic mice (18).
Recruitment of pol II.
We first compared pol II recruitment between the endogenous mouse and transgenic wild-type human β-globin loci. Recruitment to the human β-globin promoter was approximately 50% of the mouse βmaj-globin promoter results, and recruitment to the mouse 5′HS2 core was approximately 15% of that of the βmaj-globin promoter results. A similar ratio was detected at the human locus; the binding of pol II at the human 5′HS2 core was also 15% of the human β-globin promoter results.
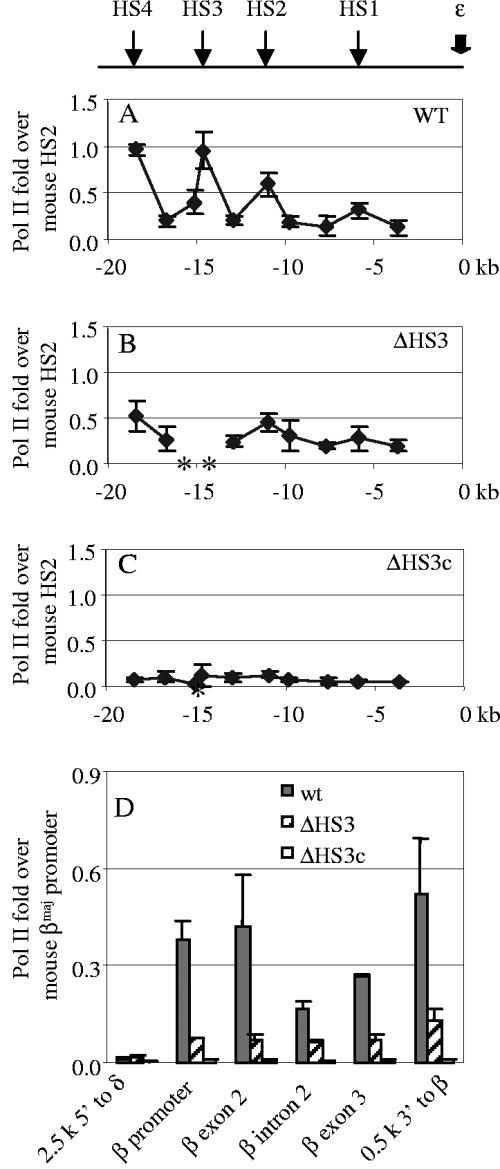
Figure 3A to C shows the distribution of pol II recruitment to the LCR in transgenic mice carrying the different βYAC constructs. In the wild-type βYAC mice, binding of pol II peaked in the HS1, -2, -3, and -4 cores, and lower levels of pol II binding were detected in the regions between them (Fig. 3A). This profile is similar to the distribution of histone acetylation in the LCR and suggests that the core sequences are likely responsible for the recruitment of pol II in addition to histone acetyltransferases. The 2.3-kb 5′HS3 deletion produced a mild reduction of pol II recruitment, but the distribution profile was the same as that seen with the wild-type βYAC mice (Fig. 3B). The 234-bp 5′HS3 core deletion eliminated pol II recruitment; only a background level of pol II binding was detected in the LCR (Fig. 3C).
FIG. 3.
pol II recruitment in the LCR and the β gene. (A) pol II recruitment in the LCR region in the wild-type (WT) βYAC mice. The positions of HSs of the LCR are shown in the top line diagram. The numbers labeled on the x axis are sequence coordinates, with zero set at the canonical cap site of the ɛ gene. The asterisks on the x axis indicate the deleted sequences in the mutant constructs. The y axis shows increases of pol II compared with that in the mouse HS2 core. Standard variation is shown as small vertical bars at each measurement point. (B) pol II recruitment in the LCR region in the 2.3-kb HS3 deletion βYAC mice. (C) pol II recruitment in the LCR region in the 234-bp HS3 core deletion βYAC mice. (D) pol II recruitment in the human β gene region. The measured positions of pol II recruitment are shown on the x axis. The y axis shows increases of pol II compared with that in the mouse βmaj promoter. wt, wild type.
Figure 3D shows the recruitment of pol II in the β-globin gene. We measured five regions of the β-globin gene: the promoter, exon 2, intron 2, exon 3, and the 3′ enhancer. pol II was highly recruited in all regions, including intron 2 in the wild-type βYAC transgenic mice. The relatively even distribution of pol II recruitment across the β-globin gene contrast to that at the LCR, but it was expected given that pol II molecules processively track the entire gene during transcription. The 2.3-kb 5′HS3 deletion considerably reduced pol II recruitment at the β-globin gene, but distribution of pol II binding was evenly maintained across the gene (Fig. 3D). Notably, the reduction of pol II binding at the β-globin gene promoter was not proportional to the decrease of the β-globin gene expression: a fourfold decrease in the pol II binding was measured, whereas β-globin gene expression was only decreased by 25% to 30%. The 234-bp 5′HS3 core deletion abolished pol II binding throughout the β-globin gene region similarly to its effect on histone acetylation (Fig. 3D). The positive correlation between the level of expression and pol II loading on the LCR compared to the promoter results implies that the LCR-bound pol II might be the enzyme actually used in transcription (22-24, 28, 60). This notion is in agreement with the observations that the pol II recruited into the LCR is in the C-terminal domain-phosphorylated form, which is associated with transcription elongation (22, 48, 60).
Recruitment of pol II was undetectable for all βYAC constructs in the region 2.5 kb 5′ to the δ-globin gene promoter, where a putative intergenic transcription promoter has been proposed to be located (Fig. 3D).
Recruitment of GATA1.
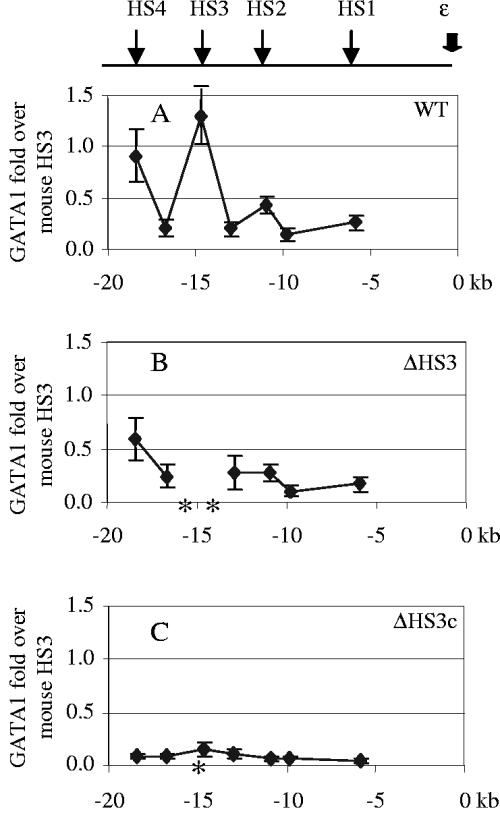
The profile of GATA1 recruitment to the LCR was similar to that of pol II (Fig. 4). GATA1 binding peaked at the LCR 5′HS cores, with a very low level of binding at the sequences between the cores. In wild-type βYAC mice GATA1 binding at the cores of 5′HSs 4 and 3 was 90% and 130% of that of the mouse 5′HS3 core, respectively, and it was 30 to 40% at the HSs 1 and 2 cores, respectively, compared to the mouse 5′HS3 (Fig. 4A). The 2.3-kb 5′HS3 deletion resulted in a general decrease of GATA1 recruitment throughout the LCR. For instance, binding of GATA1 at the 5′HS4 core was reduced from 90% to 60% in these mice (Fig. 4B). The 234-bp 5′HS3 core deletion abolished GATA1 recruitment to the LCR (Fig. 4C). A small amount of GATA1 was detected at the human β-globin gene promoter in wild-type βYAC mice, but the level was 30% of that at the mouse 5′HS3 (data not shown).
FIG. 4.
GATA-1 recruitment in the LCR. (A) GATA-1 recruitment in the wild-type (WT) βYAC mice. (B) GATA-1 recruitment in the 2.3-kb HS3 deletion βYAC mice. (C) GATA-1 recruitment in the 234-bp HS3 core deletion βYAC mice. The positions of HSs of the LCR are shown in the upper line diagram. The numbers labeled on the x axis are sequence coordinates, with zero set at the canonical cap site of the ɛ gene. The asterisks on the x axis indicate the deleted sequences in the mutant constructs. The y axis shows increases of GATA-1 compared with that seen with the mouse HS3 core. Standard variation is shown as small vertical bars at each measurement point.
Recruitment of NF-E2.
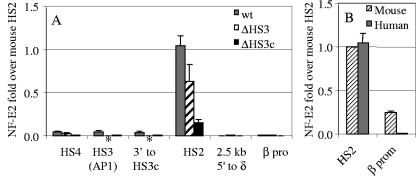
The profile of NF-E2 recruitment was different from those of pol II and GATA1. In wild-type βYAC transgenic mice, recruitment of NF-E2 at the human 5′HS2 core reached a level as high as that of the mouse 5′HS2 (Fig. 5B). In comparison with 5′HS2, the level of recruitment was only 5% at 5′HS3 and -4 (Fig. 5A), although both sites contain a NF-E2 binding motif. The 2.3-kb 5′HS3 deletion resulted in a 30% decrease of NF-E2 recruitment to 5′HS2, and the 234-bp HS3 core deletion reduced it to 15% of that of the wild-type control.
FIG. 5.
NF-E2 recruitment in the LCR and the β gene promoter. (A) NF-E2 recruitment in the LCR region. The measured positions of NF-E2 recruitment are indicated on the x axis. The asterisks on the x axis indicate the deleted sequences in the mutant constructs. The y axis shows increases of NF-E2 compared with that in the mouse HS2 core. Standard variation is shown as small vertical bars at each measurement point. wt, wild type; β pro, β gene promoter. (B) Comparison of NF-E2 recruitment on the human and mouse HS2 cores and between the promoters of the human β gene and of the mouse βmaj gene. β prom, β gene promoter.
NF-E2 recruitment at the human β-globin gene promoter was less than 1% of that at 5′HS2 in the wild-type βYAC mice (Fig. 5B). A further reduction was not observed in either the 2.3-kb 5′HS3 or 234-bp 5′HS3 core deletion mice. In contrast to the human locus, NF-E2 was highly recruited to the mouse βmaj-globin promoter; the level was 24% of that at the 5′HS2 core (Fig. 5B). This difference could be attributed to the promoter sequences; there is an imperfect NF-E2 motif in the downstream βmaj promoter (one mismatch to the consensus sequence), and NF-E2 binding on this motif can be detected when the gene is activated in MEL cells (25, 29). On the other hand, the corresponding site in the human β gene promoter is defective (four mismatches to the consensus sequence).
DISCUSSION
The holocomplex.
Results in this study support the hypothesis that the LCR forms a holocomplex and that it acts as a single entity (6, 7, 62). Deletion of either 2.3-kb 5′HS3 or 234-bp 5′HS3 core negatively affects all cis-acting elements that comprise the LCR. Although the severity of the effects is different for the two mutants, each mutation exerts its impact on histone acetylation, HS formation, and recruitment of pol II, GATA1, or NF-E2 at all measured sites. At first glance, the global effect revealed in this study contradicts the finding showing that a single 5′HS knockout (2 to 3 kb) does not impair formation of the remaining HS (2). In fact, both studies show that the formation of the remaining HSs in the context of a large deletion is detectable. In addition to this, our study demonstrated that full formation of HSs was decreased by the deletions, particularly with the 5′HS3 core deletion, which severely impaired or abolished the formation of most HSs. This global effect implies that the LCR is organized into a unified global architecture.
The impact of the two deletion mutations extends through to the δ- and β-globin genes that are 50 kb downstream of the deleted 5′HS3 site. Histone acetylation in the δ- and β-globin gene region is reduced or eliminated by the mutants. This result is consistent with the study showing that deletion of 5′HS2 to -5 in a human β locus contained in a MEL hybrid cell bearing human chromosome 11 disrupted histone acetylation in the δ- and β-globin gene region (49). Together, these studies suggest that the interconnected structure in the β-globin locus is not limited to the LCR but also includes the globin genes. However, in LCR knockout mice histone acetylation at the mouse βmaj-globin promoter is at a level similar to that observed in wild-type mice (50). Thus, the absence of the LCR has different effects on the human and mouse loci.
The LCR-coordinated domain.
The impact of 5′HS3 mutations is broad; their effects are not limited to the LCR and the downstream globin genes. The 234-bp 5′HS3 core mutation severely impaired or abolishes HS formation not only in the LCR but also at 5′HS6 (but not 5′HS7), approximately 28 kb 5′ to the ɛ-globin gene. 5′HS5, which is located between 5′HS6 and 5′HS3, is a chromatin insulator (33, 34, 55, 61, 63). However, 5′HS5 is unable to limit the impact of the 5′HS3 mutations on chromatin structure to the LCR. The effect obviously extends beyond the β-globin gene, since histone acetylation and factor recruitment alterations can be detected 3′ to the β-globin gene. However, 3′HS1 was absent in wild-type βYAC mice, so we were not able to assess the extent of 5′HS3 deletion-mediated changes. Nevertheless, these results suggest that LCR influence extends at least to 5′HS6 at the 5′ end of the construct and somewhere downstream to the β-globin gene.
Based on our analyses we propose that the human β-globin gene cluster is regulated at three hierarchic levels. The first level is the LCR holocomplex containing 5′HSs 1 to 5. The holocomplex is characterized by heavily acetylated histones with lightly acetylated regions at either end (references 5 and 16 and this study); in addition, it recruits pol II and other trans-acting factors (references 22, 23, 48, and 60 and this study), which form a highly concentrated pool (see below). Formation of the HS sites in this domain is independent of globin gene transcription (14, 15, 58). The second level is the 60-kb domain from 5′HS5 through the β-globin gene, which primarily causes the interaction between the LCR and the globin genes. The 60-kb region is necessary and sufficient for proper regulation of the globin genes during development in transgenic mice (26, 43, 45, 52). Sequences outside of the 60-kb domain are involved in the third level of regulation. These may be required for formation of the active chromatin hub (56) which also involves the LCR (41).
Putative structure of the LCR holocomplex.
Our data support the model of the LCR as a structurally integrated holocomplex (6, 7, 62). Based on the data reported here, we can describe the holocomplex structure with greater molecular specificity. First, only the HS core sequences contain highly acetylated histones and the intervening sequences between are lightly acetylated (16, 60), suggesting that acetylation of histones does not spread very far beyond each of the HS cores. Second, the impact of deleting either the whole 5′HS3 site or only the 5′HS3 core is global and proportional, suggesting that the LCR has a single unified structure. It has been suggested that chromatin with heavily acetylated histones is highly flexible (20, 31, 57; Q. Li et al., unpublished data). Here, we propose that the HS cores, due to their higher acetylation and associated flexibility in erythroid cells, are able to fold in close contact with one another, with the stiffer intervening sequences looped out between them (Fig. 6A). Because each HS core contains multiple protein binding sites, this structure allows efficient recruitment of trans-acting factors into a geometrically small space termed the LCR core. This high concentration of transcription factors bound to the LCR core catalyzes transcription activation. Since pol II is recruited at the HS cores, but not in between, this distribution suggests that if the LCR sequences are transcribed, the transcription cannot be processive and produce a long transcript. The LCR core with highly concentrated pol II in it (22, 24, 30) may be equivalent to the transcription factories that have been visualized in nuclei (8, 40). If transcription of the globin genes occurs within the LCR core, then only γ-globin gene or β-globin gene could be transcribed at a given time (62).
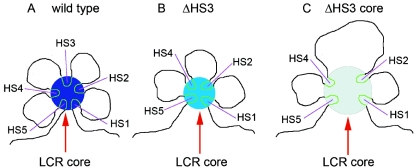
FIG. 6.
A cartoon of putative chromatin architecture of the LCR holocomplex. (A) Wild type. Chromatin encompassing the HS cores (∼200 to ∼300 bp) of HSs 1 to 5 loops together, forming the LCR core (dark-blue circle), whereas the sequences between the cores (∼3 to ∼4 kb) are looped out. Since the cores contain multiple binding motifs of trans factors, this structure results in a high concentration of trans factors in a small region (dark-blue circle), which constitutes the biochemical condition to facilitate a highly efficient transcription. (B) The 2.3-kb HS3-deleted deletion. The deletion will not considerably change length and stiffness of the chromatin between HS2 and HS4. Thus, the LCR core is retained in a similar dimension. The medium-blue circle represents the reduced concentration of trans factors due to the lack of contribution from the HS3 core. (C) The 234-bp HS3 core deletion. When the 0.2-kb HS3 core is deleted, the distance between HS 2 and HS4 is increased to ∼7 kb. The longer chromatin, which is stiffer than the HS cores, hinders a closer folding of the remaining HS cores, resulting in an increase of dimension of the LCR core. This leads to a decrease by a cubic factor in the concentration of trans factors (light-blue circle).
When a 2- to 3-kb segment of a 5′HS site is deleted, the length of the intervening sequence between the adjacent remaining HSs is roughly maintained, as is chromatin flexibility of the HS cores (Fig. 6B). Thus, the remaining HS cores are constrained together as described above. If each core sequence (5′HSs 1 to 5) contributes equally to the recruitment, then the concentration of trans-acting factors in the LCR core will be reduced to approximately four-fifths by deletion one of the HS cores. This will produce a mild decrease in β-globin gene expression, provided that the pol II recruited in the LCR core is responsible for transcription efficiency. This prediction of the additive function of the HSs of the LCR is consistent with the results for mice with 2- to 3-kb single HS knockouts (3, 4, 13, 21) and for transgenic mice carrying the 2- to 3-kb single 5′HS deletions (36, 38, 41, 42).
When the 200- to 300-bp core sequence of individual HS sites is deleted, the physical length of the intervening sequence between the two adjacent HS sites is doubled, and the longer and stiffer intervening sequences restrict chromatin bending between the adjacent cores. Thus, the HS core deletions will result in an increase in the spatial diameter of the LCR core (Fig. 6C). Since sphere volume is proportional to the cube of radius, doubling the diameter of the LCR core would increase the volume by eightfold (23); protein concentration would then be decreased by the same factor. As assumed above, deletion of a single HS core would reduce protein concentration to four/fifths of the original level. Therefore, if the diameter increases 2-fold, the concentration of trans factors in the LCR core will decrease 10-fold (4/5 × 1/23); if the diameter increases 3-fold, the concentration will decrease ∼34-fold (4/5 × 1/33). Obviously, this magnitude of trans-acting factor concentration decrease will have a catastrophic effect on globin gene expression (6, 7, 37, 38, 41). As mentioned previously, the 5′HS3 core deletion resulted in an increase in position susceptibility of β gene expression in adult, whereas no position effects on γ gene expression were observed in the early developmental stage. In this study, we analyzed a line in which β gene expression was the lowest of the all established transgenic lines (38). Although we did not perform similar analysis of other lines, it is possible that the holocomplex formed in the 5′HS3 core-less LCR might be substantially susceptible to the chromatin structure surrounding integration sites.
Both synergistic and additive stimulation of globin gene transcription by the LCR has been demonstrated in many carefully designed studies (3, 4, 6, 7, 13, 21, 36-38, 41, 42). These two apparently contradictory properties (reviewed in reference 10) can be reconciled by the LCR core architecture described above. However, this putative holocomplex structure is oversimplified and will need to be refined. For instance, the functions of each HS site are not equal and interchangeable (6, 7, 54). In addition, the holocomplex structure in embryonic erythroid cells may not be identical to that of the adult (41). Nevertheless, the concept that the HS chromatin forms a LCR catalytic center (22, 24, 30) in adult erythroid cells provides a starting point for experimentally testing the holocomplex structure.
Acknowledgments
We thank Grainne Barkess for helpful discussion and critically reading the manuscript.
This work was supported by National Institutes of Health grants DK61805 and HL73439 (to Q.L.), DK45365 (to G.S.), and DK61804 (to K.R.P.).
REFERENCES
- 1.Bender, M. A., M. Bulger, J. Close, and M. Groudine. 2000. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell 5:387-393. [DOI] [PubMed] [Google Scholar]
- 2.Bender, M. A., M. G. Mehaffey, A. Telling, B. Hug, T. J. Ley, M. Groudine, and S. Fiering. 2000. Independent formation of DnaseI hypersensitive sites in the murine beta-globin locus control region. Blood 95:3600-3604. [PubMed] [Google Scholar]
- 3.Bender, M. A., A. Reik, J. Close, A. Telling, E. Epner, S. Fiering, R. Hardison, and M. Groudine. 1998. Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse beta-globin locus control region. Blood 92:4394-4403. [PubMed] [Google Scholar]
- 4.Bender, M. A., J. N. Roach, J. Halow, J. Close, R. Alami, E. E. Bouhassira, M. Groudine, and S. N. Fiering. 2001. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood 98:2022-2027. [DOI] [PubMed] [Google Scholar]
- 5.Bulger, M., D. Schubeler, M. A. Bender, J. Hamilton, C. M. Farrell, R. C. Hardison, and M. Groudine. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell. Biol. 23:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bungert, J., U. Dave, K. C. Lim, K. H. Lieuw, J. A. Shavit, Q. Liu, and J. D. Engel. 1995. Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 9:3083-3096. [DOI] [PubMed] [Google Scholar]
- 7.Bungert, J., K. Tanimoto, S. Patel, Q. Liu, M. Fear, and J. D. Engel. 1999. Hypersensitive site 2 specifies a unique function within the human beta-globin locus control region to stimulate globin gene transcription. Mol. Cell. Biol. 19:3062-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, P. R. 1999. The organization of replication and transcription. Science 284:1790-1795. [DOI] [PubMed] [Google Scholar]
- 9.Duan, Z. J., X. Fang, A. Rohde, H. Han, G. Stamatoyannopoulos, and Q. Li. 2002. Developmental specificity of recruitment of TBP to the TATA box of the human gamma-globin gene. Proc. Natl. Acad. Sci. USA 99:5509-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel, J. D. 2001. Simple math for the beta-globin locus control region. Blood 98:2000. [Google Scholar]
- 11.Epner, E., A. Reik, D. Cimbora, A. Telling, M. A. Bender, S. Fiering, T. Enver, D. I. Martin, M. Kennedy, G. Keller, and M. Groudine. 1998. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol. Cell 2:447-455. [DOI] [PubMed] [Google Scholar]
- 12.Farrell, C. M., A. Grinberg, S. P. Huang, D. Chen, J. G. Pichel, H. Westphal, and G. Felsenfeld. 2000. A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse beta-globin locus. Proc. Natl. Acad. Sci. USA 97:14554-14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiering, S., E. Epner, K. Robinson, Y. Zhuang, A. Telling, M. Hu, D. I. Martin, T. Enver, T. J. Ley, and M. Groudine. 1995. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 9:2203-2213. [DOI] [PubMed] [Google Scholar]
- 14.Forrester, W. C., S. Takegawa, T. Papayannopoulou, G. Stamatoyannopoulos, and M. Groudine. 1987. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 15:10159-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester, W. C., C. Thompson, J. T. Elder, and M. Groudine. 1986. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc. Natl. Acad. Sci. USA 83:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser, P., J. Hurst, P. Collis, and F. Grosveld. 1990. DNaseI hypersensitive sites 1, 2 and 3 of the human beta-globin dominant control region direct position-independent expression. Nucleic Acids Res. 18:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosveld, F., G. B. van Assendelft, D. R. Greaves, and G. Kollias. 1987. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975-985. [DOI] [PubMed] [Google Scholar]
- 19.Hardison, R., J. L. Slightom, D. L. Gumucio, M. Goodman, N. Stojanovic, and W. Miller. 1997. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205:73-94. [DOI] [PubMed] [Google Scholar]
- 20.Horn, P. J., and C. L. Peterson. 2002. Chromatin higher order folding: wrapping up transcription. Science 297:1824-1827. [DOI] [PubMed] [Google Scholar]
- 21.Hug, B. A., R. L. Wesselschmidt, S. Fiering, M. A. Bender, E. Epner, M. Groudine, and T. J. Ley. 1996. Analysis of mice containing a targeted deletion of β-globin locus control region 5′ hypersensitive site 3. Mol. Cell. Biol. 16:2906-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, K. D., J. A. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 23:6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, S. H., K. Vieira, and J. Bungert. 2002. Combining chromatin immunoprecipitation and DNA footprinting: a novel method to analyze protein-DNA interactions in vivo. Nucleic Acids Res. 30:e44. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufman, R. M., C. T. Pham, and T. J. Ley. 1999. Transgenic analysis of a 100-kb human beta-globin cluster-containing DNA fragment propagated as a bacterial artificial chromosome. Blood 94:3178-3184. [PubMed] [Google Scholar]
- 27.Kulozik, A. E., S. Bail, A. Bellan-Koch, C. R. Bartram, E. Kohne, and E. Kleihauer. 1991. The proximal element of the beta globin locus control region is not functionally required in vivo. J. Clin. Investig. 87:2142-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leach, K. M., K. Nightingale, K. Igarashi, P. P. Levings, J. D. Engel, P. B. Becker, and J. Bungert. 2001. Reconstitution of human beta-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol. Cell. Biol. 21:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leach, K. M., K. F. Vieira, S. H. Kang, A. Aslanian, M. Teichmann, R. G. Roeder, and J. Bungert. 2003. Characterization of the human beta-globin downstream promoter region. Nucleic Acids Res. 31:1292-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levings, P. P., and J. Bungert. 2002. The human beta-globin locus control region. Eur. J. Biochem. 269:1589-1599. [DOI] [PubMed] [Google Scholar]
- 31.Li, Q., H. Han, X. Ye, M. Stafford, G. Barkess, and G. Stamatoyannopoulos. 2004. Transcriptional potentials of the beta-like globin genes at different developmental stages in transgenic mice and hemoglobin switching. Blood Cells Mol. Dis. 33:318-325. [DOI] [PubMed] [Google Scholar]
- 32.Li, Q., K. R. Peterson, X. Fang, and G. Stamatoyannopoulos. 2002. Locus control regions. Blood 100:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Q., and G. Stamatoyannopoulos. 1994. Hypersensitive site 5 of the human beta locus control region functions as a chromatin insulator. Blood 84:1399-1401. [PubMed] [Google Scholar]
- 34.Li, Q., M. Zhang, H. Han, A. Rohde, and G. Stamatoyannopoulos. 2002. Evidence that DNase I hypersensitive site 5 of the human beta-globin locus control region functions as a chromosomal insulator in transgenic mice. Nucleic Acids Res. 30:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Q. L., B. Zhou, P. Powers, T. Enver, and G. Stamatoyannopoulos. 1990. Beta-globin locus activation regions: conservation of organization, structure, and function. Proc. Natl. Acad. Sci. USA 87:8207-8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milot, E., J. Strouboulis, T. Trimborn, M. Wijgerde, E. de Boer, A. Langeveld, K. Tan-Un, W. Vergeer, N. Yannoutsos, F. Grosveld, and P. Fraser. 1996. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87:105-114. [DOI] [PubMed] [Google Scholar]
- 37.Navas, P. A., K. R. Peterson, Q. Li, M. McArthur, and G. Stamatoyannopoulos. 2001. The 5′HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J. Mol. Biol. 312:17-26. [DOI] [PubMed] [Google Scholar]
- 38.Navas, P. A., K. R. Peterson, Q. Li, E. Skarpidi, A. Rohde, S. E. Shaw, C. H. Clegg, H. Asano, and G. Stamatoyannopoulos. 1998. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol. Cell. Biol. 18:4188-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ney, P. A., B. P. Sorrentino, K. T. McDonagh, and A. W. Nienhuis. 1990. Tandem AP-1-binding sites within the human beta-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev. 4:993-1006. [DOI] [PubMed] [Google Scholar]
- 40.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 41.Patrinos, G. P., M. de Krom, E. de Boer, A. Langeveld, A. M. Imam, J. Strouboulis, W. de Laat, and F. G. Grosveld. 2004. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 18:1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson, K. R., C. H. Clegg, P. A. Navas, E. J. Norton, T. G. Kimbrough, and G. Stamatoyannopoulos. 1996. Effect of deletion of 5′HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc. Natl. Acad. Sci. USA 93:6605-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson, K. R., G. Zitnik, C. Huxley, C. H. Lowrey, A. Gnirke, K. A. Leppig, T. Papayannopoulou, and G. Stamatoyannopoulos. 1993. Use of yeast artificial chromosomes (YACs) for studying control of gene expression: correct regulation of the genes of a human beta-globin locus YAC following transfer to mouse erythroleukemia cell lines. Proc. Natl. Acad. Sci. USA 90:11207-11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philipsen, S., D. Talbot, P. Fraser, and F. Grosveld. 1990. The beta-globin dominant control region: hypersensitive site 2. EMBO J. 9:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porcu, S., M. Kitamura, E. Witkowska, Z. Zhang, A. Mutero, C. Lin, J. Chang, and K. M. Gaensler. 1997. The human beta globin locus introduced by YAC transfer exhibits a specific and reproducible pattern of developmental regulation in transgenic mice. Blood 90:4602-4609. [PubMed] [Google Scholar]
- 46.Reik, A., A. Telling, G. Zitnik, D. Cimbora, E. Epner, and M. Groudine. 1998. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol. 18:5992-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan, T. M., R. R. Behringer, N. C. Martin, T. M. Townes, R. D. Palmiter, and R. L. Brinster. 1989. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human beta-globin gene expression in transgenic mice. Genes Dev. 3:314-323. [DOI] [PubMed] [Google Scholar]
- 48.Sawado, T., J. Halow, M. A. Bender, and M. Groudine. 2003. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 17:1009-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 50.Schubeler, D., M. Groudine, and M. A. Bender. 2001. The murine beta-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc. Natl. Acad. Sci. USA 98:11432-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatoyannopoulos, G., and F. Grosveld. 2001. Hemoglobin switching, p. 135-182. In G. Stamatoyannopoulos, P. W. Majerus, R. M. Perlmutter, and H. Varmus (ed.), Molecular basis of blood diseases, 3rd ed. W. B. Saunders Publishing Company, Philadelphia, PA.
- 52.Strouboulis, J., N. Dillon, and F. Grosveld. 1992. Developmental regulation of a complete 70-kb human beta-globin locus in transgenic mice. Genes Dev. 6:1857-1864. [DOI] [PubMed] [Google Scholar]
- 53.Talbot, D., S. Philipsen, P. Fraser, and F. Grosveld. 1990. Detailed analysis of the site 3 region of the human beta-globin dominant control region. EMBO J. 9:2169-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanimoto, K., Q. Liu, J. Bungert, and J. D. Engel. 1999. The polyoma virus enhancer cannot substitute for DNase I core hypersensitive sites 2-4 in the human beta-globin LCR. Nucleic Acids Res. 27:3130-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanimoto, K., A. Sugiura, A. Omori, G. Felsenfeld, J. D. Engel, and A. Fukamizu. 2003. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol. Cell. Biol. 23:8946-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 57.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tuan, D., W. Solomon, Q. Li, and I. M. London. 1985. The “beta-like-globin” gene domain in human erythroid cells. Proc. Natl. Acad. Sci. USA 82:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuan, D. Y., W. B. Solomon, I. M. London, and D. P. Lee. 1989. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “beta-like globin” genes. Proc. Natl. Acad. Sci. USA 86:2554-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vieira, K. F., P. P. Levings, M. A. Hill, V. J. Crusselle, S. H. Kang, J. D. Engel, and J. Bungert. 2004. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J. Biol. Chem. 279:50350-50357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wai, A. W., N. Gillemans, S. Raguz-Bolognesi, S. Pruzina, G. Zafarana, D. Meijer, S. Philipsen, and F. Grosveld. 2003. HS5 of the human beta-globin locus control region: a developmental stage-specific border in erythroid cells. EMBO J. 22:4489-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wijgerde, M., F. Grosveld, and P. Fraser. 1995. Transcription complex stability and chromatin dynamics in vivo. Nature 377:209-213. [DOI] [PubMed] [Google Scholar]
- 63.Yu, J., J. H. Bock, J. L. Slightom, and B. Villeponteau. 1994. A 5′ beta-globin matrix-attachment region and the polyoma enhancer together confer position-independent transcription. Gene 139:139-145. [DOI] [PubMed] [Google Scholar]