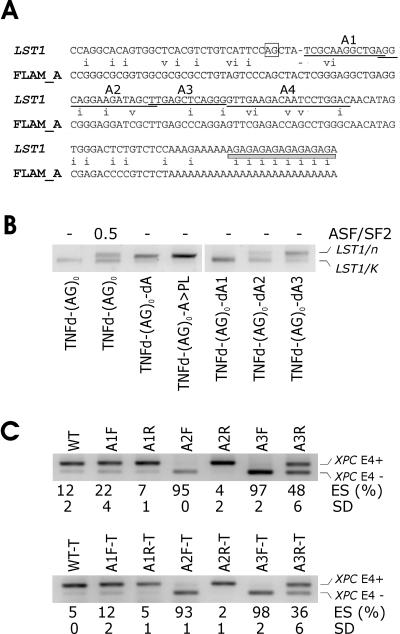
FIG. 2.
Identification of Alu-derived splicing signals that inhibit LST1/n. (A) Cryptic 3′ ss and segment A sequences are derived from Alu repeats. Sequence alignment was generated by the RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). The cryptic 3′ ss (position 33/34 in the Alu consensus sequence) (2, 37) is boxed. Deleted segments A1 (positions 40 to 51 in the Alu consensus), A2 (positions 51 to 66), A3 (positions 66 to 77), and A4 (positions 78 to 96) are underlined. The telomeric end of TNFd is shown as a gray bar. Mismatches are indicated by the letter i (transition) or v (transversion). (B) Activation of cryptic 3′ ss upon removal of segment A3 or A2. The amount of plasmid expressing ASF/SF2 is in micrograms. Mutated constructs (bottom) are described in the text. d, deletion; >, replacement. PL refers to a polylinker sequence. Expression of LST1/C and LST1/O was negligible and is not shown. Reporter constructs containing the TNFd repeat that lacked segment A were not obtained because of a detection bias against RNA isoforms with this STR (see Fig. 1B and Discussion). (C) A3 and A2 in the sense orientation promote skipping of XPC exon 4. (Upper panel) Mutated minigenes transfected into 293T cells are shown at the top. F and R, forward (sense) and reverse (antisense) orientation, respectively. ES, exon skipping as a ratio of transcripts lacking exon 4 (E4−) to the sum of E4− and E4+ transcripts. SD, standard deviation as calculated from two transfection experiments. (Lower panel) ES following transfection of the truncated (T) XPC minigene carrying identical insertions.