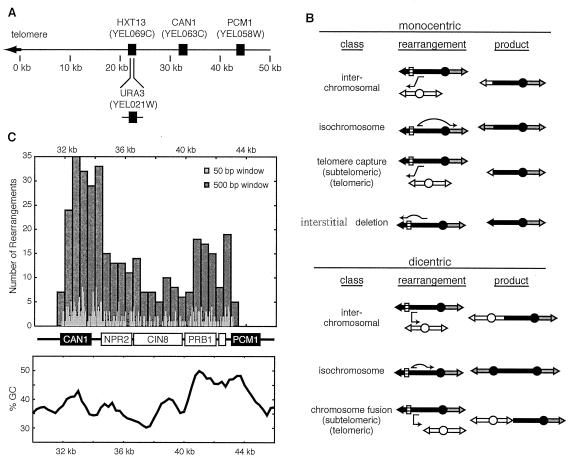
FIG. 1.
Rearrangements selected by the GCR assay can target multiple chromosomal sites. (A) Positions of the genes important to the GCR assay on the left arm of chromosome V. HXT13 was replaced by URA3. Rearrangements are selected by simultaneous counterselection against CAN1 and URA3. (B) The GCRs recovered included a variety of nonreciprocal translocations that generate mono- and dicentric products. Chromosome fusions result from dicentric translocations into telomeres or subtelomeric repeats of other chromosomes. (C) Distribution of breakpoints on chromosome V for the GCR assay. The breakpoints for all GCR isolates are plotted along the 15-kb region between the most centromeric marker (CAN1) and the most telomeric essential gene (PCM1). The 358 rearrangements are displayed at a 50 (light gray)- and a 500 (dark gray)-nucleotide resolution. The relative GC content of the chromosomal arm, calculated for a sliding 100-bp window, is displayed below the genes.