Abstract
A subset of proteins targeted by the N-end rule pathway bear degradation signals called N-degrons, whose determinants include destabilizing N-terminal residues. Our previous work identified mouse UBR1 and UBR2 as E3 ubiquitin ligases that recognize N-degrons. Such E3s are called N-recognins. We report here that while double-mutant UBR1−/− UBR2−/− mice die as early embryos, the rescued UBR1−/− UBR2−/− fibroblasts still retain the N-end rule pathway, albeit of lower activity than that of wild-type fibroblasts. An affinity assay for proteins that bind to destabilizing N-terminal residues has identified, in addition to UBR1 and UBR2, a huge (570 kDa) mouse protein, termed UBR4, and also the 300-kDa UBR5, a previously characterized mammalian E3 known as EDD/hHYD. UBR1, UBR2, UBR4, and UBR5 shared a ∼70-amino-acid zinc finger-like domain termed the UBR box. The mammalian genome encodes at least seven UBR box-containing proteins, which we propose to call UBR1 to UBR7. UBR1−/− UBR2−/− fibroblasts that have been made deficient in UBR4 as well (through RNA interference) were significantly impaired in the degradation of N-end rule substrates such as the Sindbis virus RNA polymerase nsP4 (bearing N-terminal Tyr) and the human immunodeficiency virus type 1 integrase (bearing N-terminal Phe). Our results establish the UBR box family as a unique class of E3 proteins that recognize N-degrons or structurally related determinants for ubiquitin-dependent proteolysis and perhaps other processes as well.
Protein degradation by the ubiquitin (Ub) system controls the levels of many intracellular proteins. A substrate of the Ub system is conjugated to Ub through the action of three enzymes, E1, E2, and E3 (13, 23, 30, 59, 60). A ubiquitylated protein bears a covalently linked poly-Ub chain and is degraded by the 26S proteasome (7, 63, 83). The selectivity of ubiquitylation is determined mainly by E3, which recognizes a substrate's degradation signal (degron) (2, 3, 21, 30, 48, 60). The term Ub ligase denotes either an E2-E3 holoenzyme or its E3 component. The E3 proteins are an exceptionally large family, with more than 500 distinct E3s in a mammal (13, 59, 60). An essential determinant of one class of degrons, called N-degrons, is a substrate's destabilizing N-terminal residue. The set of destabilizing residues in a given cell type yields a rule, called the N-end rule, which relates the in vivo half-life of a protein to the identity of its N-terminal residue (2, 46, 78). An N-degron consists of two major determinants: a destabilizing N-terminal residue of a protein substrate and its internal Lys residue(s) (71, 78). The latter is the site of formation of a substrate-linked poly-Ub chain (14, 61).
The N-end rule has a hierarchic structure (Fig. 1A). Specifically, N-terminal Asn and Gln are tertiary destabilizing residues in that they function through their deamidation, by N-terminal amidohydrolases (5, 41), to yield the secondary destabilizing N-terminal residues Asp and Glu (Fig. 1A). The activity of Asp and Glu requires their conjugation, by ATE1-encoded isoforms of Arg-tRNA-protein transferase (R-transferase), to Arg, one of the primary destabilizing residues (42, 43). The latter are recognized by E3 Ub ligases of the N-end rule pathway, which target a substrate for Ub-dependent degradation by the 26S proteasome (22, 46, 47, 74, 78, 85) (Fig. 1A). In mammals, the set of N-end rule destabilizing residues that function through their arginylation contains not only Asp and Glu but also Cys, which is a stabilizing (unarginylated) residue in the yeast Saccharomyces cerevisiae (3, 25, 42). The arginylation of N-terminal Cys is preceded by its oxidation (42) (R. G. Hu, J. Sheng, X. Qi, Z. Xu, T. Takahashi, and A. Varshavsky, unpublished data).
FIG. 1.
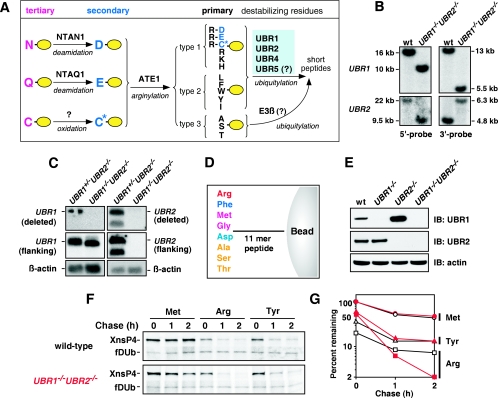
Construction and analysis of UBR1−/− UBR2−/− cells. (A) The mammalian N-end rule pathway. N-terminal residues are indicated by single-letter abbreviations for amino acids. The ovals denote the rest of a protein substrate. (B) Southern analysis of UBR1 and UBR2 using wild-type (wt) and UBR1−/− UBR2−/− embryos with cDNA probes (46, 47). To determine UBR1 genotypes, DNAs from EFs were digested with SphI (5′ probe) or BamHI (3′ probe). To determine UBR2 genotypes, embryonic DNAs were digested with NheI (5′ probe) or BamHI/SalI (3′ probe). (C) Northern analysis of UBR1 and UBR2 using total RNAs from UBR1+/− UBR2−/− and UBR1−/− UBR2−/− EFs. Different UBR1 and UBR2 cDNA fragments were used to probe the deleted regions in the knockout alleles (deleted) or both the deleted regions and its flanking regions (flanking) as described (46, 47). The two different UBR2 probes detected no significant UBR2 mRNA in UBR1−/− UBR2−/− EFs, while the UBR1 probe, which encompasses both the deleted region and its 3′-flanking region, detected UBR1-specific transcripts in UBR1−/− UBR2−/− EFs. These aberrant transcripts appear to be the abnormally spliced RNAs transcribed from the PUBR1 promoter. (D) Design of bead-conjugated synthetic peptides bearing different N-terminal amino acids. N-terminal residues of bead-conjugated peptide are indicated by three-letter abbreviations. An 11-mer peptide was covalently linked to a bead as described in Materials and Methods. (E) EF cytoplasmic proteins captured by a bead-conjugated Phe-peptide were separated in SDS-PAGE (5% Tris-glycine gel) and analyzed by immunoblotting (IB) using anti-UBR1 and UBR2 antibodies. The bottom panel shows immunoblotting analysis of actin for an input control. (F) Primary wild-type and UBR1−/− UBR2−/− EF cells were immortalized to increase transfection efficiency, and the resulting immortalized cells were transiently transfected with pcDNA3flagDHFRhaUb-X-nsP4flag (where X is Met, Arg, or Tyr) expressing fDHFRh-UbR48-X-nsP4f, a UPR-based fusion yielding cotranslationally the fDHFRh-UbR48 reference protein (denoted fDUb on the left) and X-nsP4f test protein (denoted X-nsP4 on the left). Cells were labeled for 12 min with [35S]methionine/cysteine, followed by a chase for 0, 1, and 2 h in the presence of cycloheximide, preparation of extracts, immunoprecipitation with anti-Flag M2-agarose (Sigma), SDS-PAGE, autoradiography, and quantitation, essentially as described (46). (G) Quantitation of the patterns in F using PhosphorImager. The amounts of 35S in an X-nsP4f, relative to 35S in the fDHFRh-UbR48 reference protein at the same time points, were plotted as percentages of this ratio for Met-nsP4f (bearing a stabilizing N-terminal residue) at time zero. Open and solid symbols denote data obtained with wild-type and UBR1−/− UBR2−/− extracts, respectively. ○, Met-nsP4; ▵, Tyr-nsP4; □, Arg-nsP4.
The N-end rule pathway is present in all eukaryotes examined so far, from animals and plants to fungi. It is also present in prokaryotes, despite their lacking both Ub conjugation and Ub itself (25, 46, 70, 73, 78). The known functions of the N-end rule pathway include the control of peptide import (through conditional degradation of the import's repressor) (22, 74), the fidelity of chromosome segregation (through degradation of a conditionally produced cohesin's fragment) (62), the regulation of apoptosis (through degradation of a caspase-processed inhibitor of apoptosis) (20, 77), as well as regulation of meiosis in yeasts and metazoans (6, 38, 46), leaf senescence in plants (89), and cardiovascular development in mammals (42). The pathway’s function in cardiovascular development is likely to be mediated, at least in part, by the arginylation-dependent degradation of RGS4, RGS5, and RGS16, a set of GTPase-activating proteins that down-regulate signaling by specific G proteins and are themselves targeted by the N-end rule pathway (17) (M. J. Lee, T. Tasaki, J. Y. An, K. Moroi, I. Vavydov, and Y. T. Kwon, unpublished data; R. G. Hu, J. Sheng, X. Qi, Z. Xu, T. Takahashi, and A. Varshavsky, unpublished data).
Other substrates of the N-end rule pathway (in addition to those mentioned above) include RNA polymerases of alphaviruses (18); the integrase of the human immunodeficiency virus type 1 (HIV-1) (56); a protein called p60, secreted by the bacterium Listeria monocytogenes into the host cell's cytosol (68); the 3C protease of encephalomyocarditis virus (49); and a subset of γ2 subunits of mammalian G proteins (27). The functions of the N-end rule pathway in controlling the levels of these proteins remain to be understood. In addition, the results of studies in which dipeptides with destabilizing N-terminal residues were used to perturb the N-end rule pathway suggested its involvement in cell differentiation (32, 58), turnover of muscle proteins (69), and limb regeneration in amphibians (72).
The E3 Ub ligases that recognize N-degrons are called N-recognins (6, 78). UBR1, the S. cerevisiae N-recognin, is a 225-kDa RING-type E3 containing at least three substrate-binding sites. The type 1 site binds to basic N-terminal residues (Arg, Lys, and His) of either protein-sized N-end rule substrates or short peptides. The type 2 site binds to bulky hydrophobic N-terminal residues (Leu, Ile, Phe, Tyr, and Trp) (6, 78). The third binding site of S. cerevisiae UBR1 targets substrates through their internal (non-N-terminal) degrons, and is allosterically “activated” through perturbation of autoinhibited UBR1 conformation by peptides with destabilizing N-terminal residues that bind to UBR1 at its type 1 and type 2 sites (22, 74). The only known substrate of the yeast N-end rule pathway that is targeted through the third binding site of UBR1 is CUP9, a transcriptional repressor that down-regulates PTR2 (encoding a peptide transporter) and several other genes as well (22, 74).
Our previous studies identified and characterized mammalian E3s termed UBR1 and UBR2 (45-47), two sequelogs of S. cerevisiae UBR1 (sequelog and spalog are evolutionarily neutral terms denoting a sequence that is similar to a specified extent to another sequence and a three-dimensional structure that is similar to a specified extent to another three-dimensional structure, respectively [79]). UBR2 was also characterized by another group (40). The mouse UBR1 and UBR2 are functionally overlapping 200-kDa N-recognins with 47% identity and 68% similarity to each other. Their binding specificities to destabilizing N-terminal residues of proteins or short peptides are indistinguishable from each other in nonquantitative assays (46, 47). Human UBR1 and UBR2 in HeLa cells were found to interact with RECQL4, a putative helicase that is mutated in patients with the Rothmund-Thomson and RAPADILINO syndromes, a set of recessively inherited illnesses whose multiple manifestations include predisposition to cancer (88). The currently characterized interaction of RECQL4 with UBR1 and UBR2 does not confer a short half-life on RECQL4 (88).
Previously constructed UBR1−/− mice were viable but different from their wild-type littermates in several ways that remain to be understood mechanistically (47). By contrast, UBR2−/− mice, in some of the strain backgrounds examined, exhibited gender-dependent lethality in that most UBR2−/− females died during embryogenesis, while UBR2−/− males were viable but infertile owing to postnatal apoptosis of spermatocytes (46).
In the present work, we further addressed the functions of UBR1 and UBR2 by constructing double-mutant UBR1−/− UBR2−/− mouse strains. These mice, in contrast to their single-mutant counterparts, died as early embryos. We also characterized the N-end rule pathway in fibroblasts derived from UBR1−/− UBR2−/− embryos and found that these cells, despite lacking the pathway's known Ub ligases, retained a significant fraction of the pathway's activity. We then employed an affinity assay and proteomic methods to search for other mouse N-recognins, identifying an exceptionally large (570-kDa) protein, termed UBR4, and the 300-kDa UBR5, a previously characterized mammalian E3 known as EDD/hHYD (11, 29, 31, 65). Strong sequelogs (79) of mammalian UBR4 are known as PUSHOVER in Drosophila melanogaster and as BIG in plants (19, 24, 37, 52, 54, 64, 87). RNA interference (RNAi) was used to make UBR1−/− UBR2−/− fibroblasts also deficient in UBR4. The resulting cells were impaired in the degradation of N-end rule substrates. The recognition of destabilizing N-terminal residues by UBR4 and UBR5 is a new property of these proteins. UBR1, UBR2, UBR4, UBR5, and at least three other mouse proteins contain a distinct motif that we propose to call the UBR box. The resulting UBR box family consists of either demonstrated or inferred ubiquitin ligases. A major part of this family are E3s of the N-end rule pathway.
MATERIALS AND METHODS
Construction of UBR1−/− UBR2−/− mouse strains and isolation of UBR1−/− UBR2−/− fibroblasts.
Mouse UBR1 and UBR2 are located on chromosomes 2 and 17, respectively (45, 46). UBR1+/− and UBR2+/− mice, in the 129SvImJ/C57BL/6 genetic background, were crossed to produce compound-heterozygous UBR1+/− UBR2+/− mice. The latter were intercrossed to produce, among other genotypes, the UBR1−/− UBR2−/− progeny. Construction and characterization of UBR1−/− and UBR2−/− mice was described (46, 47).
Primary embryonic fibroblasts (EFs) were established from E11.5 UBR1−/− UBR2−/− embryos and their UBR1+/− UBR2+/+ littermates, as described (46, 47). Permanent cell lines were established from primary EFs through crisis-mediated immortalization, by replating cell cultures over ∼2 months (∼1.5 × 106 cells onto a 10-cm plate every 3 days). DNA transfection efficiency was considerably higher with immortalized EFs than with primary EFs.
Antibodies.
Chicken polyclonal antibodies to mouse UBR4 were produced by Gallus Immunotech Inc. (Ontario, Canada) against synthetic peptides CSFEKYDEDHSGDDK (residues 4615 to 4629; antibody UBR4-119R) and DLDGEDEKDKGALDC (residues 2982 to 2995, plus an additional C-terminal Cys; antibody UBR4-244). Rabbit (peptide-mediated) polyclonal antibodies to mouse UBR1 and UBR2 were produced and characterized as described (46, 47). Mouse monoclonal antibodies to N-terminal and C-terminal regions of HIV-1 integrase were a gift from Dag Helland (University of Bergen, Norway). Anti-V5 epitope antibody was purchased from Invitrogen (Carlsbad, CA). Rabbit antibodies to mouse UBR5/EDD/hHYD and to green fluorescent protein (GFP) were from Santa Cruz Biotechnology (Santa Cruz, CA) and Abcam (Cambridge, MA), respectively. Antibody to Drosophila Hyperplastic disks (dHYD/dUBR5) was a gift from Allen Shearn (Johns Hopkins University, Baltimore, MD). Rabbit antiactin antibody was from Sigma. Anti-Ub antibody (Biomol International, Plymouth Meeting, PA) binds to both mono- and polyubiquitylated proteins, but not to free Ub. Secondary antibodies were from Bio-Rad (Hercules, CA) and Santa Cruz Biotechnology.
Northern hybridization analysis.
Mouse adult tissue RNA blot (Seegene, Seoul, Korea) was hybridized with 32P-labeled cDNA fragments of mouse UBR4 or human β-actin. The UBR4 cDNA probe (1,269 bp) was amplified by PCR, using the primers 5′-CACCATGGTTCCCACCTTAGGCTCAC (forward) and 5′-TGTGCGGGCTGTGATGGTAGCTG (reverse).
In vivo protein degradation assay and plasmids.
Immortalized EF cells were transiently transfected, using Lipofectamine 2000 (Invitrogen), with plasmids expressing test substrates. About 24 h after transfection, cells were labeled for 10 min at 37°C with [35S]methionine/cysteine (35S-Express, Perkin-Elmer, Boston, MA), followed by a chase in the presence of cycloheximide, preparation of extracts, immunoprecipitation, NuPAGE 10%-bis-Tris sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (Invitrogen), autoradiography, and quantitation of 35S by Molecular Imager FX system (Bio-Rad), essentially as described (41, 43).
To determine in vivo decay curves of X-nsP4 (where X is Met, Arg, Tyr, His, Phe, Gly, Leu, or Thr), cells were transfected with plasmids expressing X-nsP4 from the cytomegalovirus promoter (PCMV), with X-nsP4 as part of the fDHFRh-UbR48-X-nsP4f fusion (superscripts h and f denote the hemagglutinin and Flag epitopes, respectively), a UPR (Ub/protein/reference) construct (47). This fusion, fDHFRh-UbR48-X-nsP4f, is cotranslationally cleaved by deubiquitylating enzymes at the UbR48-X junction, yielding the long-lived fDHFRh-UbR48 reference protein and X-nsP4f, a test protein. In the UPR technique, the reference protein serves as an internal control for levels of expression, immunoprecipitation yields, sample volumes, and other sources of sample-to-sample variation (46, 51, 71, 74, 75, 80). fDHFRh-UbR48-X-nsP4f plasmids were constructed by subcloning PCR-produced SmaI-XbaI fragments encoding X-nsP4f into the EheI- and XbaI-cut UPR vector pcDNA3(dEheI)FDHUMCM (J. Sheng and A. Varshavsky, unpublished data) encoding fDHFRh-UbR48. Parameters of X-nsP4 degradation in EF cells were calculated as described (51).
The set of pEGFP-IRES-Ub-X-integrase (where X is Met, Gly, Arg, His, Leu, or Phe) plasmids expressing the enhanced green fluorescent protein (EGFP) reference protein and X-integrase (see Fig. 7A) were described previously (56). The pcDNA3-Ub-X-INMOD-IRES-hrGFP (where X is Met and Phe) plasmids were constructed by ligating the Ub open reading frame (ORF) to a synthetic version of the HIV-1 integrase gene (AF422697; Met or Phe at position 1 of integrase) and then aligning an internal ribosome entry site element with the humanized recombinant Renilla leniformis green fluorescent protein ORF (Stratagene, La Jolla, CA) such that a single transcriptional unit was specified.
FIG. 7.
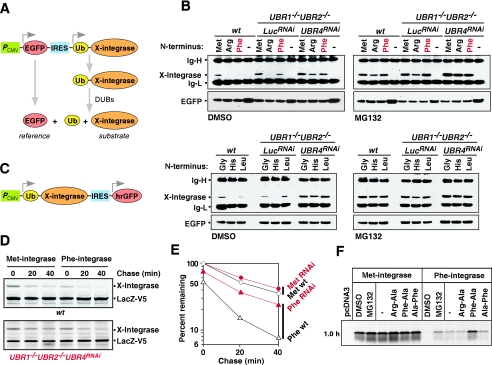
HIV-1 integrase is an in vivo substrate of UBR box proteins. (A) Schematic diagram for a bicistronic mRNA producing EGFP and X-integrase (where X is Met, Gly, Arg, His, Phe, or Leu) used in B. (B) The steady-state level of X-integrase and EGFP, expressed in wild-type, UBR1−/− UBR2−/− LucRNAi, and UBR1−/− UBR2−/− UBR4RNAi cells, was determined using immunoprecipitation and Western analysis with anti-integrase antibody. Cells were treated with MG132 (10 μM) or solvent (dimethyl sulfoxide [DMSO]). N-terminal amino acids of integrase are indicated on the top. Ig-H and Ig-L, mouse immunoglobulin G heavy and light chains, respectively. The expression of the reference protein EGFP was determined using immunoblotting with anti-GFP antibody for an input control. (C) A pcDNA3-based bicistronic construct expressing Ub-X-integrase and hrGFP used in D to F. Since hrGFP was poorly expressed in EFs, LacZ-V5 was coexpressed as a reference protein. (D) Pulse-chase analysis of Met- and Phe-integrases in wild-type and UBR1−/− UBR2−/− UBR4RNAi cells. Cells were labeled for 30 min with [35S]methionine/cysteine, followed by a chase for 0, 20, and 40 min in the presence of cycloheximide, preparation of extracts, immunoprecipitation with a combination of anti-integrase and anti-V5 antibodies, followed by SDS-PAGE and autoradiography. (E) Quantitation of the patterns shown in D using a PhosphorImager. (F) Degradation of Met- and Phe-integrases was analyzed using in vitro transcription/translation-coupled rabbit reticulocyte lysate in the presence of MG132 (5 μM) or dipeptide (2 mM) as indicated, and [35S]methionine-labeled proteins were analyzed by SDS-PAGE, followed by autoradiography. Met-integrase is indeed slightly stabilized by MG132 or the type 1 dipeptide Arg-Ala (but not by the type 1 dipeptide Phe-Ala), suggesting that a minor population of Met-integrase may be modified (e.g., oxidation) into an unknown structure that mimics a weak type 1 N-degron.
To examine steady-state levels of X-integrase, cells were transfected with one of the above plasmids and incubated for 1.5 h (∼12 h after transfection) in the presence or absence of the proteasome inhibitor MG132 (EMD Biosciences, La Jolla, CA). Cells were lysed in 1% Nonidet-P40 (NP-40), 0.15 M NaCl, 1 mM EDTA, 10 mM Tris-HCl (pH 7.5), and the lysates were incubated with monoclonal antibody to HIV-1 integrase. Immunocomplexes were recovered using protein G-agarose (Amersham Bioscience, Piscataway, NJ) and analyzed using NuPAGE 10%-Bis-Tris gel (Invitrogen), followed by immunoblotting with the same antibody. Lysates were also subjected to immunoblotting with anti-GFP antibody. For pulse-chase analysis of X-integrase, plasmid pcDNA3.1D/V5-His/lacZ (Invitrogen) encoding β-galactosidase-V5-His was cotransfected to normalize the level of 35S-labeled X-integrase.
In vitro transcription-translation-degradation assay.
The test protein was expressed in 25 μl of TNT Quick-coupled transcription-translation system (Promega, Madison, WI) containing a rabbit reticulocyte lysate premixed with most of the reaction components necessary to carry out transcription/translation in the lysate. X-integrase was labeled with [35S]methionine (Amersham Bioscience). A 2-μl aliquot of the reaction was taken out at each time point and mixed with 8 μl of SDS sample loading buffer, followed by SDS-PAGE and autoradiography. X-nsP4 was labeled with biotin by adding biotinylated lysine-tRNA complex in the reaction (Transcend tRNA, Promega). To detect biotinylated X-nsP4, the blotted membrane was incubated with horseradish peroxidase-conjugated streptavidin (Pierce), followed by enhanced chemiluminescence. The reaction mixtures containing dipeptides also contained 0.15 mM bestatin (Sigma), an inhibitor of some aminopeptidases, to reduce degradation of the added dipeptides (47).
Peptide-based pulldown assay.
For affinity-based identification of mouse N-recognins, peptides X-Ile-Phe-Ser-Thr-Asp-Thr-Gly-Pro-Gly-Gly-Cys (where X is Arg, Phe, Gly, Ser, Thr, Ala, Asp, or Met) were synthesized (Fig. 1D). The sequence of residues 2 to 9 of these peptides was identical to that of Sindbis virus polymerase (nsP4) at the same positions. The peptide was cross-linked, via its C-terminal Cys residue, to UltraLink Iodoacetyl beads (Pierce, Rockford, IL), as described (22). Testes and brains were collected from 2- to 6-month-old male C57BL6/J mice (Charles River, Wilmington, MA) and frozen immediately in liquid N2. The tissues were homogenized in buffer A (10% glycerol, 75 mM KCl, 0.1 mM EDTA, 20 mM HEPES, pH 7.9) containing protease inhibitors (Sigma), and a soluble fraction was prepared by a two-step centrifugation, at 9,000 × g for 20 min and at 100,000 × g for 1 h.
Cytoplasmic extracts of EFs and D.Mel-2 cell (ATCC CRL 1963) were prepared as described (88). Extracts were diluted to 2 mg ml− 1 of protein in buffer B (0.05% Tween 20, 10% glycerol, 0.2 M KCl, 20 mM HEPES, pH 7.9). In some experiments with cytoplasmic extracts, buffer C (0.1% NP-40, 10% glycerol, 0.15 M KCl, 20 mM HEPES, pH 7.9) was used instead of buffer B. Peptide-linked beads (15-μl packed volume) were incubated in 1 ml of tissue or cytoplasmic extracts diluted with buffer B (2 mg of total protein) at 4°C for 12 h. The beads were pelleted by centrifugation in a microcentrifuge at 2,500 rpm for 30 seconds, washed four times with 0.5 ml of buffer B, resuspended in 70 μl of SDS-PAGE sample buffer containing 0.1 M dithiothreitol, and heated at 70°C for 10 min. For analysis by mass spectrometry, the beads were heated at 56°C for 30 min. For elution with dipeptides, the above washed (by buffer B) beads were washed again with buffer D (75 mM NaCl, 20 mM HEPES, pH 7.9) and incubated with 0.25 ml of 25 mM dipeptide (Phe-Ala or Ala-Phe; Bachem, King of Prussia, PA) for 10 min on ice. After pelleting the beads, the supernatants were pooled and concentrated using Microcon YM-10 (Millipore, Bedford, MA).
Mass spectrometry.
Silver staining of SDS-PAGE gels was performed according to an online protocol (http://www.healthsystem.virginia.edu/internet/biomolec/Silverstains.cfm). For mass spectrometry, separated proteins were blotted onto a polyvinylidene difluoride membrane (Problott, Applied Biosystems, Foster City, CA) and stained using the colloidal gold method (Bio-Rad). The immobilized proteins were reduced, S-carboxymethylated, and digested in situ with Achromobacter protease I (Lys-C), which cleaves after Lys (33). Mass spectrometry analyses of protein fragments were performed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry using a PerSeptive Biosystem Voyager-Delayed Extraction/RP (35). Proteins were identified by comparing molecular masses determined by mass spectrometry with molecular masses derived from sequence databases.
RNAi.
RNAi was carried out essentially as described (9). Four 19-nucleotide sequences were chosen from the mouse UBR4 ORF, using the Whitehead siRNA Selection Web Server (http://jura.wi.mit.edu/bioc/siRNA) (91). Four pairs of 64-nucleotide oligonucleotides including a 9-nucleotide hairpin spacer (5′-TTCAAGAGA-3′) were synthesized (MWG Biotech, High Point, NC), annealed, and subcloned into a BglII- and HindIII-cleaved retroviral vector pSUPER.retro.puro (OligoEngine, Seattle, WA). The target sequences used were A (nucleotides 202 to 224, 5′-GCAGTACGAGCCGTTCTA-3′), B (nucleotides 5083 to 5105, 5′-GGTATGCCATAAGGACCAT-3′), C (nucleotides 8149 to 8171, 5′-CAAGCGGAGACACGTGACG-3′), and D (nucleotides 13690 to 13712, 5′-GGATCAACTGGTGATGCTC-3′), yielding plasmids pSUPER-A to pSUPER-D, respectively. A control plasmid (pSUPER-Luc) was constructed using a 19-nucleotide sequence (5′-GATTATGTCCGGTTATGTA-3′) to target the firefly luciferase (76).
Ecotropic retroviruses for transduction of mouse cells were obtained by transient transfection of the retroviral vectors into the packaging cell line Phoenix Eco (American Type Culture Collection SD3444). Phoenix Eco cells were grown to ∼50% confluence in 10-cm culture dishes and then transfected with 4 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen). The medium was replaced 24 h after transfection; the retrovirus-containing medium was collected at 48 h after transfection and filtered through a 0.45-μm filter. Wild-type and UBR1−/− UBR2−/− EFs were plated on a 10-cm culture dish at 2 × 105 to 3 × 105 cells 12 to 18 h prior to infection. The medium was replaced with 4 ml of the infection cocktail, including 2 ml of retrovirus-containing medium, 4 μg ml− 1 of Polybrene, and a standard medium, followed by incubation for 4 h. A 7-ml aliquot of standard medium was then added to the cultures, followed by incubation for 44 h. The infected cells were split onto a new 10-cm dish and cell lines containing integrated retrovirus were selected with puromycin at 4 to 6 μg ml− 1 (Sigma).
DNA/protein sequences and alignment analysis.
Sequence alignment analysis was carried out using the T-COFFEE web server (http://www.ch.embnet.org/software/TCoffee.html) (57). Phylogenetic analyses were performed using MEGA version 2.1 (39).
RESULTS
Construction and characterization of UBR1−/− UBR2−/− mouse strains and cell lines.
The retention of the N-end rule pathway in the previously produced single-mutant UBR1−/− and UBR2−/− EF cell lines was consistent with the fact that both UBR1 and UBR2 are N-recognins and thus functionally complement one another (46, 47). In the present work, we used compound heterozygous crosses of UBR1+/− UBR2+/− mice to produce double-mutant UBR1−/− UBR2−/− strains, whose genotype and the absence of wild-type UBR1 and UBR2 mRNAs were verified by Southern and Northern analyses, respectively (Fig. 1B and C). In contrast to the viability of UBR1−/− mice and (gender-biased) late-embryonic lethality of UBR2−/− mice (see the introduction), the absence of both UBR1 and UBR2 resulted in early embryonic lethality. Specifically, no live UBR1−/− UBR2−/− embryos were recovered beyond E11.5, and the recovered double-mutant embryos exhibited multiple morphological and growth abnormalities, including defective proliferation of neuroepithelial cells and apparent cardiovascular defects (J. Y. An., T. Tasaki, A. Varshavsky, and Y. T. Kwon, unpublished data).
To characterize the N-end rule pathway in the absence of both UBR1 and UBR2 and to produce UBR1−/− UBR2−/− cells that could be readily transfected with DNA, we established EF cell lines from fibroblasts of E11.5 UBR1−/− UBR2−/− embryos. A previously described, peptide-based pulldown assay (Fig. 1D) (22, 46) was used to detect either UBR1 or UBR2 in cell extracts through their binding to destabilizing N-terminal residues of proteins or short peptides. This assay utilized a set of otherwise identical 12-mer peptides with different N-terminal residues. Each peptide was cross-linked to microbeads through its C-terminal Cys residue. Immobilized X-peptides, with X being a varying N-terminal residue, were incubated with cell extracts, followed by recovery of bound proteins, SDS-PAGE, and immunoblotting with anti-UBR1 or anti-UBR2 antibodies (46). The Phe-peptide assay (Phe is a type 2 primary destabilizing residue) (Fig. 1A) confirmed that UBR1−/− UBR2−/− EFs lacked both UBR1 and UBR2, that UBR1−/− EFs lacked UBR1 but not UBR2, and that UBR2−/− EFs lacked UBR2 but not UBR1 (Fig. 1E).
Previous work identified the 70-kDa Sindbis virus RNA polymerase (nsP4) as an N-end rule substrate that is targeted for degradation, at least in rabbit reticulocyte extract, through the N-terminal Tyr residue of nsP4 (18). (In vivo, Tyr-bearing nsP4 is produced through its excision, by a viral protease, from a polyprotein synthesized during Sindbis virus infection.) X-nsP4 proteins, produced in vivo using the Ub fusion technique, are either short-lived or metabolically stable, depending on the presence of a destabilizing or stabilizing N-terminal residue, respectively (46, 47). We carried out pulse-chase assays with UBR1−/− UBR2−/− versus wild-type EF cells that had been transiently transfected with plasmids expressing fDHFRh-UbR48-X-nsP4f fusion proteins, where X was a varied residue. These reporters are based on the UPR technique (46, 47, 71, 74, 80). The long-lived reference moiety fDHFRh-UbR48, which serves as a reference protein, is cotranslationally cleaved from a nascent fDHFRh-UbR48-X-nsP4f fusion by deubiquitylating enzymes. Remarkably, despite the absence of both UBR1 and UBR2, the only two mouse N-recognins known at the time of analysis, UBR1−/− UBR2−/− EFs, were found to contain the N-end rule pathway. Its activity was detectably but not considerably decreased in UBR1−/− UBR2−/− EFs relative to their wild-type counterparts (Fig. 1F and G), suggesting that the mammalian genome encodes an unknown N-recognin that recognizes the N-degrons for Ub-dependent proteolysis.
Identification of UBR4 and UBR5/EDD, which can bind to type 1 and/or type 2 N-degrons.
To identify such an E3, we asked whether there is an additional UBR1/2-like E3. We identified a previously uncharacterized UBR-homologous protein, named UBR3, whose complete ORF is currently not available in the database. We cloned the 8-kb full-length mouse UBR3 cDNA by conventional cDNA library screening (T. Tasaki, Y. T. Kwon, and A. Varshavsky, unpublished data). The 213-kDa UBR3 is a RING-finger E3, shares weak but detectable homology (22% identity) to UBR1 and UBR2, and contains several domains conserved in UBR1 and UBR2. However, UBR3 expressed in S. cerevisiae cells did not bind to N-degrons (T. Tasaki, Y. T. Kwon, and A. Varshavsky, unpublished data), to which UBR1 and UBR2 readily bound. These results suggest that the mammalian genome encodes at least one N-recognin that does not share overall sequence similarity to UBR1 or UBR2.
We have recently developed a peptide-pulldown assay to measure the interaction between an overexpressed E3 and N-degron (Fig. 1A, D, and E) (22, 46). We then asked whether N-degron-peptide beads could precipitate the proteomic amounts of endogenous N-recognins from mouse tissue extracts. The assay employed immobilized X-peptides bearing N-terminal Arg (a type 1 destabilizing residue), Phe (a type 2 destabilizing residue), Met or Gly (stabilizing residues), Asp (an ATE1 substrate), or Ala, Ser, or Thr (type 3 destabilizing residues) (25, 78).
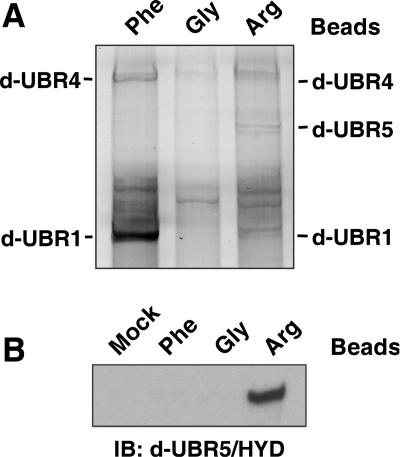
Specific X-peptides were incubated with mouse testis extracts (Fig. 2A to D) and with extracts from other tissues as well (data not shown), followed by centrifugation, isolation of peptide-bound proteins, SDS-PAGE, and silver staining. Silver staining of precipitates from peptide-based pulldown assays reproducibly revealed four major proteins that were captured by type 1 and/or type 2 peptides (Fig. 2A). Proteins blotted on a polyvinylidene difluoride membrane were visualized by the colloidal gold staining; MALDI-TOF/mass spectrometry analysis of Achromobacter protease I-digested proteins identified them as UBR1, UBR2, and two other proteins termed UBR4 and UBR5 (Fig. 2A).
FIG. 2.
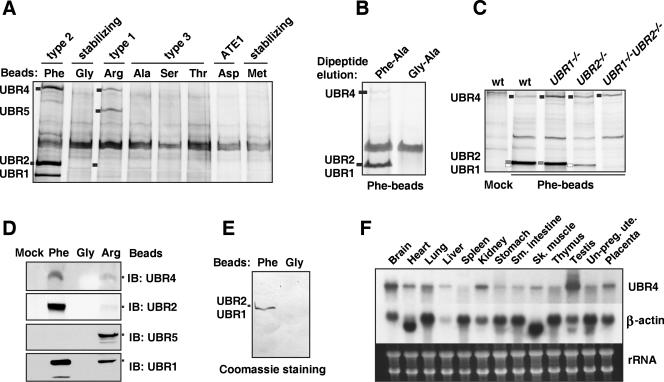
Identification of mouse N-recognins. (A) Peptide-pulldown assay using testis extracts and bead-conjugated peptides. Captured proteins were separated and visualized using silver staining. The identities of four major bands that are specifically and reproducibly captured by Phe- and/or Arg-peptide beads were determined using peptide mass fingerprinting. (B) UBR1, UBR2, and UBR4 bound from testis extract were eluted from Phe-peptide beads by the Phe-Ala dipeptide but not by Gly-Ala. (C) The amount of UBR4 bound by Phe-peptide beads from EF cell extracts does not depend strongly on the presence of UBR1 and/or UBR2. Proteins bound by either mock or Phe-peptide beads are indicated on the left with short bars: white, UBR1; gray, UBR2; and black, UBR4. (D) Testis proteins bound to bead-conjugated peptides bearing different N-terminal amino acids were immunoblotted with antibodies against UBR1, UBR2, UBR4, and UBR5. Mock, beads without peptide conjugation. (E) Semipurification of endogenous UBR1/UBR2 from an EF cytoplasmic extract using a peptide-pulldown assay. The precipitates/Phe-peptide beads complex prepared from cytoplasmic EF extracts were separated on SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, followed by staining with Coomassie brilliant blue R-250 (Bio-Rad). Anti-UBR1 and -UBR2 immunoblotting confirmed enrichment of UBR1 and UBR2 (data not shown). (F) Northern blot analysis of UBR4 and β-actin using different mouse adult tissues. Total RNA (20 μg) was loaded in each lane. Ethidium bromide-stained ribosomal RNAs are shown as a loading control (bottom panel).
The first two proteins identified were UBR1 and UBR2, which we have recently shown to bind to type 1 and type 2 N-degrons by using GST-pulldown assays with overexpressed UBR1 and UBR2 (46). Notably, endogenous UBR1 and UBR2 bound more efficiently to N-terminal Phe (a type 2 residue) than to N-terminal Arg (a type 1 residue) (Fig. 1A and 2A), suggesting that these two N-recognins have stronger affinity to type 2 than type 1 N-degrons. Quantitative binding data and functional assays are required to address the potential significance of this result.
UBR4 is a huge, novel protein, whose complete ORF is not clear in the database. Based on both the apparent size of UBR4 on SDS-PAGE (Fig. 2A) and the hypothetical 15,468-bp ORF deduced from the databases on overlapping cDNA sequences, we conjecture that the ∼109-kb mouse UBR4, on chromosome 4, comprises 108 exons and encodes an exceptionally large protein of 570 kDa. UBR4 appears to be a sequelog of the 560-kDa Arabidopsis BIG and the 560-kDa Drosophila PUSHOVER (see Discussion), given similar sizes, continuous overall homology, and shared domains, including a unique Cys-rich domain (Fig. 3B) (24, 37, 52, 54, 64, 87).
FIG. 3.
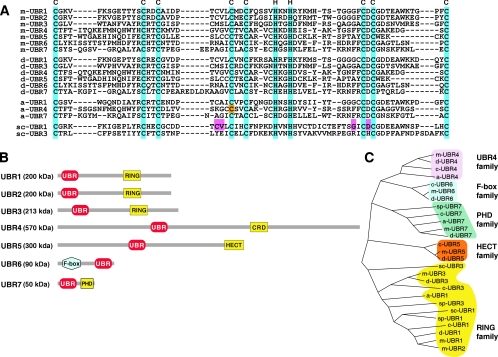
(A) Sequence alignment of the UBR box motifs. Conserved Cys and His residues are highlighted (cyan). Indicated by orange highlight is the Cys residue of Arabidopsis BIG, whose missense mutation perturbs auxin transport (24). Indicated by pink are the residues of S. cerevisiae UBR1 essential for degradation of type 1 N-end rule substrates (A. Webster, M. Ghislain, and A. Varshavsky, unpublished data). Note that different organisms contain different sets of UBR box proteins. Prefixes: m, Mus musculus; d, Drosophila melanogaster; a, Arabidopsis thaliana; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe, c, Caenorhabditis elegans. Protein sequences listed are m-UBR1 (NP_033487), m-UBR2 (AAQ17202), m-UBR3 (T. Tasaki, Y. T. Kwon, and A. Varshavsky, unpublished data), m-UBR4 (this study), m-UBR5 (AAT28194), m-UBR6 (AAH55343), m-UBR7 (BAC40212), d-UBR1 (NP_573184), d-UBR3 (NP_572426), d-UBR4 (NP_476986), d-UBR5 (P51592), d-UBR6 (AAF54459), d-UBR7 (AAF53607), c-UBR1 (AAB42328), c-UBR3 (AAO38656), c-UBR4 (AAD47122), c-UBR5 (NP_492389), c-UBR6 (AAC48052), c-UBR7 (CAA99920), a-UBR1 (NP_195851), a-UBR4 (NP_186875), a-UBR7 (T08905), sc-UBR1 (P19812), sc-UBR2 (NP_013124), sp-UBR1 (CAB10108), sp-UBR3 (O60152), and sp-UBR7 (Q09329). (B) Locations of the UBR boxes and domains characteristic, in particular, of E3 Ub ligases in the mouse UBR1 to UBR7 proteins. UBR, UBR box; RING, RING finger; CRD, cysteine-rich domain; HECT, HECT domain; PHD, plant homeodomain. (C) Phylogenetic relationships of UBR box proteins. The relative evolutional distances between each UBR box sequence were calculated by a neighbor-joining method. The tree is unrooted and the length of each branch is proportional.
Among the roles of Arabidopsis UBR4 (BIG) are regulation of the function and transport of auxin, a major plant hormone (19, 24, 37, 52, 54). In Drosophila, UBR4 (PUSHOVER) plays a role in glial cell growth, neuronal excitability, and synaptic vesicle fusion (64, 87). Northern blot analysis demonstrated that UBR4 mRNA is rather broadly expressed in mouse tissues and embryonic stages, with a higher level in the adult testis and brain (Fig. 2F). UBR1, UBR2, and UBR4 were reproducibly retained by Phe-peptide and Arg-peptide (to different extents for different UBRs; see below), but not by other X-peptides tested (Fig. 2A), suggesting that these proteins bind to both type 1 and type 2 N-degrons. A dipeptide bearing type 1 or type 2 N-degron competitively inhibits the in vitro binding of UBR1 and UBR2 to N-degron (46, 47). UBR1, UBR2 and UBR4 that were bound to the immobilized Phe-peptide could be eluted by the dipeptide Phe-Ala but not by Gly-Ala, which bore a stabilizing N-terminal residue (Fig. 2B), suggesting that UBR1, UBR2, and UBR4 may contain a common domain that binds to N-degrons. The relative amounts of UBR4 retained on Phe-peptide beads that were incubated with protein concentration-adjusted extracts from either wild-type, UBR1−/−, UBR2−/−, or UBR1−/− UBR2−/− EF cells did not differ considerably, indicating that the interaction of UBR4 with N-terminal Phe does not require UBR1/UBR2 and also suggesting that expression of UBR4 is not influenced strongly, if at all, by the level of UBR1 or UBR2 in a cell (Fig. 2C).
Another mammalian N-recognin identified by the X-peptide pulldown assay (Fig. 2A) was a 300-kDa protein called EDD (E3 identified by differential display) (11) or hHYD (31). EDD/hHYD has been shown to be an HECT-domain E3 Ub ligase essential for embryogenesis and interacting with a variety of proteins, including progesterone receptor and a topoisomerase-binding protein (29, 31, 65). This protein is a strong sequelog (79) of the previously characterized Drosophila tumor suppressor gene hyperplastic disks (55). We termed it UBR5 because its N-degron-binding property specifically classifies this E3 as a member of the UBR box proteins (see below). The UBR root is recommended as a standard root for the names of E3 Ub ligases that recognize N-degrons; see the web site (http://www.informatics.jax.org/mgihome/nomen) provided by the Jackson Laboratory, Bar Harbor, Maine. Furthermore, UBR5 (EDD/hHYD) was captured by type 1 (Arg) but not by type 2 (Phe) X-peptide beads, a binding pattern distinct from that of UBR1, UBR2, and UBR4 (Fig. 2A). Thus, UBR5 appears to be an N-recognin specific for type 1 (basic) N-terminal residues.
To further verify the results from mass spectrometry, we produced two peptide-mediated chicken antibodies to mouse UBR4. The peptide bead-bound proteins from testes extracts were eluted, separated on SDS-PAGE, and blotted with antibody against UBR1, UBR2, UBR4, or UBR5. Consistent with the silver staining data (Fig. 2A), anti-UBR1, -UBR2, and -UBR4 antibodies detected a ∼200-kDa doublet (for UBR1 and UBR2) and a 570-kDa band (for UBR4) from precipitates prepared by bead-conjugated peptides bearing a specific N-degron but not a stabilizing N-terminal residue (Fig. 2D). Anti-UBR5 antibody detected a 300-kDa band from precipitates prepared with a bead-conjugated peptide bearing type 1 N-degron but not type 2 N-degron or a stabilizing N-terminal residue (Fig. 2D). These results collectively suggest that UBR1 and UBR2 preferentially bind to type 2 N-degrons, UBR4 binds to type 1 and type 2 N-degrons, while UBR5 binds to type 1 but not to type 2 N-degrons.
Protein-based pulldown assays (e.g., immunoprecipitation or GST-pulldown assay) are usually based on affinity between specific protein sequences or conformation in a protein- or species-specific manner and thus require appropriate antibodies or DNA manipulation. In contrast, based on the universal interaction between an E3 (e.g., N-recognin) and a degron (e.g., N-degron), the peptide pulldown assay may be generally applied for one-step purification or precipitation of endogenous N-recognins (and maybe other E3s as well with specific degrons) from a broad spectrum of tissues or species (from prokaryotes to mammals) as well as for E3-substrate interaction analyses. For example, Fig. 2E shows that elution of Phe-peptide-bound proteins (captured from EF extracts; ∼2 mg total proteins) with an SDS-PAGE sample buffer yields a Coomassie quantity (0.1 to 1.0 μg) of highly enriched UBR1 and UBR2 (Fig. 2E).
UBR box family of E3 ubiquitin ligases.
The above results suggest that the mammalian genome encodes at least four proteins that can bind to type 1 and/or type 2 N-degrons. We then asked whether these proteins share a common motif involved in recognition of N-degrons. Although UBR4 and UBR5 showed no significant overall homology to other mouse proteins, UBR1 to UBR5 contained a putative ∼70-residue zinc finger-like motif named the UBR box (Fig. 3A). By using a sequence alignment program (see Materials and Methods), we found that the mammalian genome encodes at least seven of the mouse UBR box proteins termed UBR1 to UBR7 (Fig. 3A and B). Phylogenetic analysis of the UBR box motif sequences from yeast to mouse classified them into UBR4, UBR6 (with an F-box), UBR7 with the PHD (plant homeodomain) domain, UBR5 (with the HECT domain), and UBR1/2/3 (with the RING finger) subfamilies (Fig. 3C).
As described above, UBR1, UBR2, and UBR3 contain the RING ubiquitylation domain. The 300-kDa UBR5/EDD contains the HECT ubiquitylation domain and the UBA (Ub-associated) domain (11, 29). UBR6 is a 90-kDa orphan F-box protein which contains the F-box motif, a feature of degron-recognizing subunits of modular Ub ligases called SCF (13, 36, 59, 82). The F-box motif is essential for the interaction of an F-box-containing SCF subunit with the rest of SCF Ub ligase. SCFs are a large, functionally diverse family of Ub ligases that differ not only in regard to their F-box subunits but in other subunits as well (13, 59). The presence of the UBR box in UBR6 (Fig. 3A and B) and the fact that the previously characterized F-box subunits of SCF Ub ligases function as their degron-recognizing components suggest that UBR6 may recognize degrons via its UBR domain, and moreover, that the corresponding SCF E3 may function as an N-recognin, a conjecture to be tested.
UBR7 is a novel 50-kDa protein. It contains a putative PHD domain that resembles but is distinct from the RING domain. We used putative because the distinction between RING and PHD is fairly subtle, and the “true” PHD domain appears not to be present in the Ub ligases described so far (1, 12, 66). Thus, it remains to be determined whether the above domain of UBR7 is a bona fide PHD or, instead, a version of the RING domain characteristic of a large class of E3 Ub ligases that includes UBR1 and UBR2. As with the UBR6 protein above, the co-occurrence, in UBR7, of the UBR box motif and a (putative) PHD domain suggests that UBR7 is also an E3 Ub ligase, possibly an N-recognin. Based on these results, we propose that the UBR box family defines a unique E3 class that recognizes N-degrons or structurally related molecules, via the UBR box motif, for Ub-dependent proteolysis or related processes.
We also asked whether UBR box proteins that were previously known or were shown in the present work to be N-recognins also have such properties in an organism phylogenetically distant from mammals. Extracts from the Drosophila melanogaster D.Mel-2 cell line were subjected to the Phe-peptide pulldown assay, followed by SDS-PAGE of recovered proteins and silver staining. The results (Fig. 4) were in agreement with the above expectation. Specifically, the Phe-peptide assay isolated two major species whose sizes were consistent with their being Drosophila UBR1 (called UBR1 in Drosophila as well) and UBR4 (called PUSHOVER in Drosophila) (64) (Fig. 4A). The Arg-peptide assay captured, in addition, a protein of the size of Drosophila UBR5 (HYD) (Fig. 4A), a result consistent with the preferential binding of mouse UBR5 (EDD/hHYD) to Arg-peptide over Phe-peptide (Fig. 3A). Immunoblotting of the same samples with antibody to Drosophila UBR5 confirmed the recovery of Drosophila UBR5 by the Arg-peptide assay (Fig. 4B). Although preliminary, these findings suggest that strong sequelogs of mammalian UBR1, UBR2, UBR4, and UBR5 in phylogenetically distant eukaryotes also function as N-recognins, that is, the N-end rule pathway's E3 Ub ligases, or their subunits.
FIG. 4.
N-recognins in Drosophila melanogaster. (A) Peptide-pulldown analysis of Drosophila proteins in D.Mel-2 cells. Cytoplasmic proteins were precipitated by a bead-conjugated peptide bearing N-terminal Arg, Phe, or Gly. (B) Peptide-pulldown analysis followed by Western blot using anti-Drosophila UBR5/HYD antiserum.
Degradation of N-end rule substrates is impaired in cells deficient in UBR1, UBR2, and UBR4.
UBR5 (EDD/hHYD) is the previously characterized E3 Ub ligase (11, 29, 31, 65) whose specificity as an N-recognin was discovered in the present work (Fig. 2A and D). In contrast, there is no published evidence (apart from sequence comparisons) about the Ub ligase activity of either Drosophila PUSHOVER or plant BIG, the previously characterized strong sequelogs of mammalian UBR4 (11, 19, 24, 37, 52, 54, 64, 87). Both the presence of the UBR box in UBR4 (PUSHOVER/BIG) (Fig. 3A) and the demonstrated specific binding of UBR4 to destabilizing N-terminal residues (Fig. 2A to D) strongly suggest that UBR4 is an N-recognin.
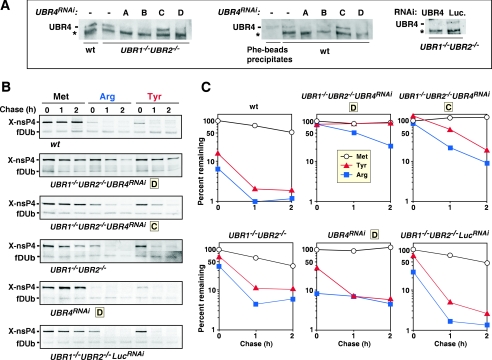
To address this issue further, we employed retrovirus-mediated RNAi to construct UBR4RNAi, UBR1−/− UBR2−/− UBR4RNAi, and UBR1−/− UBR2−/− luciferase (UBR1−/− UBR2−/− LucRNAi) stable cell lines (Fig. 5A). Among the four siRNAs tested (sequences A to D), siRNAs A, B, and D yielded a strong (80 to 90%) decrease in UBR4 protein as determined by anti-UBR4 immunoblotting (Fig. 5A), whereas siRNA C did not yield significant UBR4 silencing. We then examined the in vivo stability of 70-kDa Sindbis virus RNA polymerase Tyr-nsP4 (type 2 N-degron, an authentic protein) and Arg-nsP4 (its mutant bearing a type 1 N-degron) in wild-type and various UBR protein mutant cells by using pulse-chase analysis (Fig. 5B and C).
FIG. 5.
Analysis of the in vivo degradation of Sindbis virus RNA polymerases bearing N-terminal Tyr in wild-type, UBR4RNAi, UBR1−/− UBR2−/− UBR4RNAi, and UBR1−/− UBR2−/− LucRNAi stable cell lines. (A) Immunoblot analysis of UBR4 in UBR4RNAi, UBR1−/− UBR2−/− UBR4RNAi, and UBR1−/− UBR2−/− LucRNAi stable cell lines. Wild-type (wt) and UBR−/− UBR2−/− EFs stably expressing UBR4 short-hairpin RNA (shRNA) were analyzed by immunoblot using anti-UBR4 antibody. Cells were transduced by recombinant retroviruses with four different short-hairpin RNA sequences (A, B, C, and D) for UBR4 RNAi or firefly luciferase (Luc) and subsequently selected for puromycin resistance. After drug selection, a pool of colonies was lysed, and UBR4 knockdown efficiency was determined using 5% SDS-PAGE and immunoblotting. A nonspecific band is indicated by the asterisk as a loading control. Independent immunoblot analysis with isolated colonies gave largely similar results (data not shown). (B) Pulse-chase analysis of X-nsP4, where X is Met (stabilizing), Arg (type 1 N-degron), or Tyr (type 2 N-degron), in cells deficient in UBR proteins, singly or in combination. See Fig. 1F for details. (C) Quantitation of the patterns shown in B using PhosphorImager.
While 1.9% and 10% of Tyr-nsP4 remained after the 2-h chase in wild-type and UBR1−/− UBR2−/− cells, ∼85% of Tyr-nsP4 remained in two independent UBR1−/− UBR2−/− UBR4RNAi cell lines under the same condition (Fig. 5B and C and Table 2, and data not shown). Further, Tyr-nsP4 was rapidly ubiquitylated in reticulocyte lysates, and its ubiquitylation was specifically inhibited by a dipeptide bearing a type 2 N-degron but not a type 1 N-degron (Fig. 6C). In contrast to Tyr-nsP4, 1.2%, 6.0%, and 26% of Arg-nsP4 remained in wild-type, UBR1−/− UBR2−/−, and UBR1−/− UBR2−/− UBR4RNAi cells, respectively (Fig. 5B and C), suggesting that there is an additional N-recognin (e.g., UBR5) other than UBR1, UBR2, and UBR4 in normally growing mouse EFs.
TABLE 2.
Half-lives of X-nsP4 in wild-type and mutant EF cellsa
| N-terminal residue type | Residue | Half-life (mm)
|
|||||
|---|---|---|---|---|---|---|---|
| Wild-type EFs
|
UBR1−/− UBR2−/− EFs
|
||||||
| No treatment | UBR4RNAi D | No treatment | UBR4RNAi D | UBR4RNAi C | LucRNAi | ||
| Stabilizing | Met | 75 | >120 | 84 | >120 | >120 | 96 |
| Gly | >120 | 64 | >120 | 88 | |||
| Type 1 | Arg | 42 | >120b | 34 | 72 | 40 | 32 |
| His | 36 | 50 | >120 | 50 | |||
| Type 2 | Tyr | 40 | 37 | 38 | >120 | 56 | 32 |
| Phe | 38 | 54 | >120 | 39 | |||
| Leu | 40 | 55 | 113 | 41 | |||
| Type 3 | Thr | 61 | 52 | 99 | 48 | ||
Half-life (t0.50-60, min) of X-nsP4 between 0 and 60 min of chase was calculated as follows: t0.50-60 = 50 (%) × 60 (min)/SD0-60 (%). Subsequent decay (SD0-60, %) denotes the ratio of the amount of a labeled X-nsP4 at 60 min of chase to that of the same labeled X-nsP4 at time zero. SD0-60 = 100 − {[X-nsP4]60/[X-nsP4]0} × 100, where [X-nsP4]60 was the amount of 35S-labeled X-nsP4 at 60 min and [X-nsP4]0 was as described in Table 1, footnote a.
FIG. 6.
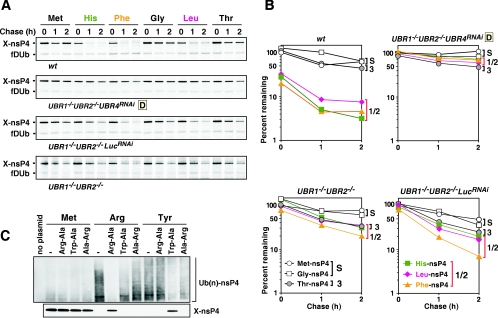
Degradation kinetics of X-nsP4 bearing N-terminal His, Phe, Gly, Leu, and Thr in various UBR mutant cells. (A) Pulse-chase analysis of X-nsp4, where X is Met or Gly (stabilizing), His (type 1 N-degron), Leu or Phe (type 2 N-degron), or Thr (type3 N-degron), in cells deficient in UBR proteins, singly or in combination. (B) Quantitation of the patterns shown in A using PhosphorImager. S, stabilizing; 1/2, type 1/2 N-degron; 3, type 3 N-degron. (C) Ubiquitylation assay of X-nsP4 in rabbit reticulocyte lysate. Upper panel: X-nsP4 proteins (where X is Met, Arg, or Tyr) were expressed and translationally labeled with biotin using transcription-translation-coupled reticulocyte lysates with or without 2 mM dipeptide as indicated. The proteasome inhibitor MG132 (5 μM) was also present, to accumulate ubiquitylated proteins. After 30 min of incubation, ubiquitylated proteins were immunoprecipitated with anti-Ub antibody, followed by SDS-PAGE of precipitated proteins and detection of biotinylated proteins (see Materials and Methods). Lower panel: X-nsP4 proteins (where X is Met, Arg, or Tyr) were in vitro translated as above without MG132 and a portion of the reticulocyte lysates was loaded, followed by Western blotting with horseradish peroxidase-conjugated streptavidin.
The degree of stabilization of Tyr-nsP4 and Arg-nsP4 was in agreement with the degree of UBR4 silencing because they remained short-lived in UBR1−/− UBR2−/− LucRNAi control cells and was only marginally stabilized in UBR1−/− UBR2−/− UBR4RNAi cell line C, where no significant UBR4 knockdown was observed (Fig. 5B and C). Further, degradation of these substrates was not significantly impaired in UBR4RNAi or UBR1−/− UBR2−/− cells, suggesting that either UBR4 knockdown or UBR1/UBR2 knockout is not sufficient to perturb nsP4 degradation.
Together with our finding that endogenous UBR4 binds to N-degrons (Fig. 2D), these results collectively implicate UBR4 as a third N-recognin and suggest that there are at least three mammalian N-recognins that can recognize type 1 and type 2 N-degrons. N-recognin is currently defined as an E3 that can recognize N-degrons for ubiquitylation. However, since it is unclear whether UBR4 has a ubiquitylation domain, an N-recognin can also be a substrate (N-degron) recognition subunit of an E3 complex.
The relative destabilizing activities of N-terminal residues have not been characterized extensively in mammalian cells. Given the N-end rule-dependent proteolysis of Tyr-nsP4, we next examined the functional interaction of UBR box proteins with other N-terminal amino acids (Fig. 6A and B and Table 2). In addition to type 1 and type 2 N-degrons, the N-terminal Ala, Ser, and Thr residues have also been found to confer metabolic instability on an N-end rule reporter (X-[β-galactosidase]) in reticulocyte extract (25), suggesting the existence of a distinct (type 3) branch of the N-end rule pathway.
As shown in Fig. 6B and Table 2, Thr-nsP4 bearing a putative type 3 destabilizing N-terminal residue was a long-lived protein in all genetic backgrounds tested, leaving the problem of type 3 N-degrons to be addressed in future work. In contrast, X-nsP4s bearing N-terminal His (type 1), Leu (type 2), or Phe (type 2) were short-lived in wild-type EF cells (Fig. 6B and Table 2). Interestingly, the degradation of His-nsP4, Leu-nsP4, and Phe-nsP4 was significantly slower in UBR1−/− UBR2−/− cells, suggesting that participation of N-recognins other than UBR1 and UBR2 in targeting the above destabilizing residues may be relatively minor. Taken together with other data above, these findings (Fig. 6A and B and Table 2) suggest that the type 1 and type 2 substrate-binding sites of UBR1, UBR2, UBR4, UBR5, and other (to be discovered and/or verified) N-recognins may recognize specific destabilizing N-terminal residues of a given type (either 1 or 2) differentially. The relative destabilizing activities of the N-terminal residues tested, with X-nsP4 as a reporter, are summarized in Tables 1 and 2.
TABLE 1.
Initial decay of X-nsP4 in wild-Type and mutant EF cellsa
| N-terminal residue type | Residue | Initial decayb of X-nsP4 (%)
|
|||||
|---|---|---|---|---|---|---|---|
| Wild-type EFs
|
UBR1−/− UBR2−/− EFs
|
||||||
| No treatment | UBR4RNAi D | No treatment | UBR4RNAi D | UBR4RNAi C | LucRNAi | ||
| Stabilizing | Met | 0 | 0 | 0 | 0 | 0 | 0 |
| Gly | 0 | 0 | 8 | 0 | |||
| Type 1 | Arg | 86c | 92 | 54c | 17 | 14 | 72 |
| His | 71 | 0 | 23 | 6 | |||
| Type 2 | Tyr | 74c | 65 | 36c | 18 | 0 | 29 |
| Phe | 80 | 23 | 32 | 19 | |||
| Leu | 67 | 4 | 16 | 0 | |||
| Type 3 | Thr | 0 | 0 | 20 | 0 | ||
Initial decay (ID12, %) of X-nsP4 at the end of a 12-min pulse was calculated as follows: ID12 = 100 − {[X-nsP4]0/[Met-nsP4]0} × 100 where [X-nsP4]0 denotes the amount of 35S-labeled X-nsP4 at time 0 (at the end of the 12-min pulse) after normalization as described for Fig. 1G.
Zero indicates that the relative amount of 35S-labeled X-nsP4 at time zero was higher than that of Met-nsP4 in the same cell line.
UBR1, UBR2, and UBR4 participate in degradation of HIV-1 integrase.
The 32-kDa integrase of human immunodeficiency virus is produced in infected cells through its excision, by HIV-encoded protease, from the 160-kDa HIV Gag-Pol polyprotein and subsequently mediates the insertion of the (reverse-transcribed) viral genome into a host's chromosome (8). We have previously shown that HIV-1 integrase (Phe-integrase), which naturally bears N-terminal Phe, a type 2 destabilizing residue, is short-lived in human HEK-293T cell line, and its N-terminal Phe is a part of an N-degron that mediates the bulk of Phe-integrase degradation in these cells (56). However, specific E3s that mediate Phe degradation have not been characterized. We examined steady-state levels of X-integrase proteins (where X is Met, Gly, Arg, His, Phe, and Leu) in wild-type, UBR1−/− UBR2−/− LucRNAi, and UBR1−/− UBR2−/− UBR4RNAi cells. In the expression system used (Fig. 7A), the EGFP (fluorescent) protein reference is expressed from the PCMV promoter, while an X-integrase reporter is expressed from an internal ribosome entry site as part of the Ub-X-integrase fusion that is cotranslationally cleaved by deubiquitylating enzymes in vivo to yield X-integrase.
In wild-type cells, the levels of X-integrase bearing either type 1 or type 2 N-terminal residues (including Phe, its natural N-terminal residue) were significantly lower than the levels of X-integrases bearing stabilizing N-terminal residues, such as Met or Gly (Fig. 7B). Furthermore, the levels of normally short-lived X-integrase proteins were significantly increased in UBR1−/− UBR2−/− UBR4RNAi cells in comparison to UBR1−/− UBR2−/− LucRNAi cells (Fig. 7B), suggesting that UBR1, UBR2, and UBR4 participate in the in vivo targeting of these integrases. However, treatment of cells with MG132, a proteasome inhibitor, significantly increased the levels of all the integrase proteins tested, including Met-integrase, even in UBR1−/− UBR2−/− UBR4RNAi cells, indicating the presence of yet another E3(s) that targets N-degrons and/or an internal degron of integrase.
We then attempted to evaluate the in vivo stability of X-integrase in wild-type and UBR protein mutant cells using pulse-chase analysis. Due to low in vivo labeling efficiency, we humanized the codons of integrase (see accession number AF422697) to increase translation efficiency and fused the resulting ORF to the C terminus of Ub to generate N-terminal Phe (Fig. 7C). When normalized to the levels of β-galactosidase (encoded by a cotransfected plasmid), the amount of 35S-labeled Phe-integrase remaining after 40 min of chase was increased from 14% in wild-type cells to 32% in UBR1−/− UBR2−/− UBR4RNAi cells (Fig. 7D and E).
To further examine the in vitro proteolysis of X-integrase (where X is Arg, Phe, or Met), X-integrase was expressed and labeled with [35S]methionine in transcription-translation-coupled rabbit reticulocyte lysates. In agreement with the above results, it was rapidly destroyed in reticulocyte extract and could be stabilized by Phe-Ala (type 2) dipeptide but not by either Arg-Ala (type 1) dipeptide or Ala-Phe (Fig. 7F). These results collectively suggest that HIV-1 Phe-integrase can be at least in part targeted through the recognition of its N-degron by a set of cellular E3 systems, including UBR1, UBR2, and UBR4.
To test whether the in vivo integration of HIV-1 into cellular DNA is impaired in cells that lack specific N-recognins, wild-type, UBR1−/− UBR2−/−, and UBR1−/− UBR2−/− UBR4RNAi cells were infected with GFP-marked recombinant HIV-1, and the integration of HIV-1 was assessed using fluorescence-activated cell sorting. The knockout or knockdown of UBR1, UBR2, and UBR4 marginally impaired HIV-1 infection (L. C. F. Mulder, T. Tasaki, Y. T. Kwon, and M. A. Muesing, unpublished data), suggesting that the inhibition of these three molecules is not sufficient to block HIV-1 integration. However, it remains to be tested whether HIV-1 infectivity is significantly decreased in human cells deficient in UBR1, UBR2, and UBR4 and if other UBR box proteins play an additional role in HIV-1 integrase turnover.
DISCUSSION
We have previously shown that two functionally overlapping mammalian E3s, termed UBR1 and UBR2, can recognize N-degrons for N-end rule-dependent proteolysis (45-47). Given the retention of this pathway in single-mutant UBR1−/− and UBR2−/− mouse cell lines (46) (see the introduction), we here constructed mice lacking both UBR1 and UBR2 (Fig. 1B and C) and demonstrate that UBR1−/− UBR2−/− cells retain significant degradation activities for model N-end rule substrates (Fig. 1F and G). This result is unexpected because the S. cerevisiae N-end rule pathway is mediated only by one N-recognin, the 225-kDa UBR1 (6).
To identify an additional N-recognin, that is, an E3 Ub ligase that recognizes N-degrons, we performed an affinity-based proteomic approach using synthetic peptides bearing N-degrons. We report here that four mammalian proteins, UBR1, UBR2, a 570-kDa novel protein termed UBR4, and a 300-kDa E3 called EDD (termed UBR5 in this study), can bind to type 1 and/or type 2 N-degrons and that the mammalian genome encodes at least seven proteins (termed UBR1 to UBR7) sharing a ∼70-amino-acid zinc finger-like domain termed the UBR box motif.
A remarkable feature of UBR box proteins is that they all (except for UBR4) contain specific body signatures (e.g., RING in UBR1, UBR2, and UBR3; HECT in UBR5; F-box in UBR6; PHD in UBR7) found in E3 or a substrate recognition subunit of an E3 complex, suggesting that the UBR box family defines a unique E3 class, most likely N-recognin. Is the UBR box motif the only signature that defines N-recogin? The relatively small (45 kDa) Arabidopsis PRT1 Ub ligase has recently been shown to act as an N-recognin that recognizes a subset of type 2 N-degrons (70). However, this RING finger E3 does not contain the “canonical” UBR box motif (Fig. 2A). Interestingly, we found that the ZZ domain of PRT1 weakly resembles a part of the UBR box motif (T. Tasaki and Y. T. Kwon, unpublished data), suggesting that variants of the UBR box motif may exist and recognize distinct subsets of N-degrons or structurally related molecules.
Data from pulse-chase analysis indicate that normally short-lived Tyr-nsP4 is almost completely stabilized in UBR1−/− UBR2−/− UBR4RNAi cells, whereas the same protein is only marginally stabilized in UBR1−/− UBR2−/− cells (Fig. 5B and C). Together with peptide pulldown assays indicating that UBR4 can bind to a type 2 N-degron (Fig. 2), in vivo degradation assays indicate that UBR4 is a third N-recognin (in addition to UBR1 and UBR2). Notably, UBR1−/− UBR2−/− UBR4RNAi cells retained a weak but detectable activity for degradation of Arg-nsP4 bearing a type 1 N-degron (Fig. 5B and C), predicting the presence of a fourth N-recognin mainly specific for type 1 substrates. Our biochemical data indicate that mouse and Drosophila UBR5 proteins can bind to a type 1 (but not type 2) N-degron (Fig. 2A and D and Fig. 4A and B), suggesting that UBR5 is likely to be a type 1-specific N-recognin responsible for the residual type 1 N-recognin activities observed in UBR1−/− UBR2−/− UBR4RNAi cells.
During proteomic screening of N-recognins, we did not detect UBR3, UBR6, or UBR7, suggesting that these UBR box proteins may not bind efficiently to N-degrons under these conditions. Consistent with this observation, mouse UBR3 expressed in S. cerevisiae did not bind to N-degrons (T. Tasaki, Y. T. Kwon, and A. Varshavsky, unpublished data) to which UBR1 and UBR2 readily bound under the same conditions (46). Nonetheless, we do not exclude the possibility that UBR6 and UBR7 may not have been expressed in normally growing mouse EFs or required a posttranslational modification (e.g., phosphorylation, sumoylation, or neddylation) to be active as N-recognins. In mammals, N-terminal Ala, Ser, and Thr have been reported to be destabilizing through their recognition by an unidentified E3 called E3β with a size of ∼200 kDa (25, 28). However, our attempts to capture such an E3 using type 3 peptide beads were unsuccessful.
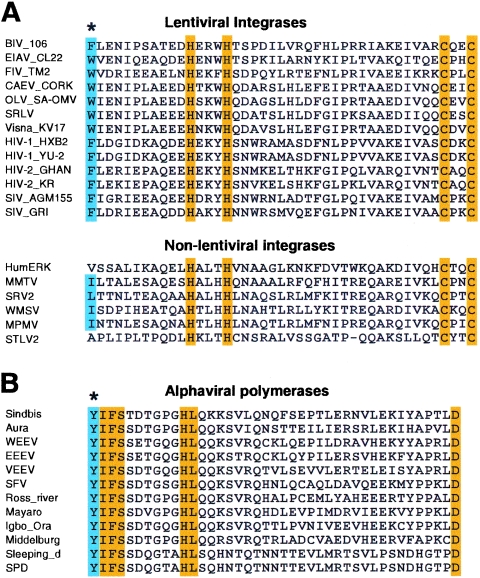
Sindbis virus RNA polymerase nsP4 and HIV-1 integrase have been suggested as substrates of the N-end rule pathway (18, 56). Our results (Fig. 5 to 7) provide the functional connection between these substrates and their cellular E3s and suggest that the N-end rule pathway may participate in degradation of a broad range of viral proteins bearing N-degrons and thus for their viral life cycle as well. Given the conserved nature of interactions between N-degrons and E3, we examined the identity of the N-terminal amino acids of lentiviral integrases (including HIV-1 integrase) and alphaviral RNA polymerases (including Sindbis polymerase). As summarized in Fig. 8, all of the viral proteins that we have examined thus far bore type 2 N-degrons, suggesting that the in vivo stabilities of these viral proteins (and thus their life cycles as well) may be regulated through their N-degron-mediated proteolysis by a set of cellular UBR box proteins.
FIG. 8.
Lentiviral integrases and alphaviral RNA polymerases commonly bear type 2 N-degron. (A, B) Alignment of inferred N-terminal sequences of mature lentiviral integrases and nonlentiviral integrases (A) and alphaviral RNA polymerases (B). Conserved type 2 N-degrons are indicated by blue highlighting and an asterisk. Other conserved residues are indicated by orange highlighting. The protein sequences (accession number, organism) listed are: BIV_106 (P19560, bovine immunodeficiency virus isolate 106), EIAV_CL22 (P32542, equine infectious anemia virus clone CL22), FIV_TM2 (P31822, feline immunodeficiency virus isolate TM2), CAEV_CORK (P33459, caprine arthritis encephalitis virus strain CORK), OLV_SA_OMV (P16901, ovine lentivirus strain SA-OMVV), SRLV, (AAS18421, Small ruminant lentivirus), visna_KV17 (P35956, Visna lentivirus strain KV1772), HIV-1_HXB2 (P04585, HIV-1 HXB2 isolate), HIV-1_YU-2 (P35963, HIV-1 YU-2 isolate), HIV-2_GHAN (P18042, HIV-2 isolate GHANA-1), HIV-2_KR (Q74120, HIV-2 isolate KR), SIV_AGM155 (P27973, simian immunodeficiency virus AGM155 isolate), SIV_GRI-1 (Q02836, simian immunodeficiency virus isolate AGM), HumERK (CAA76879, human endogenous retrovirus K), MMTV (GNMVMM, mouse mammary tumor virus), SRV2 (AAD43256, simian retrovirus 2), WMSV (NP_041261, woolly monkey sarcoma virus), MPMV (GNLJMP, Mason-Pfizer monkey virus), STLV2 (CAA74901, simian T-lymphotropic virus 2), WEEV (P_818936, western equine encephalomyelitis virus), EEEV (S72349, eastern equine encephalitis virus), VEEV (AAD14556, Venezuelan equine encephalitis virus), SFV (NP_463457, Semliki forest virus), Mayaro (NP_579968, Mayaro virus), Sindbis (P03317, Sindbis virus), Sleeping_d (NP_740656, sleeping disease virus), Igbo_Ora (NP_740715, Igbo Ora virus), SPD (NP_740638, salmon pancreas disease virus), Aura (NP_819013, aura virus), Middelburg (P03318, Middleburg virus), and Ross_river (P13887, Ross River virus strain NB5092).
Mammalian UBR box proteins (UBR1 to UBR7) have their apparent functional homologs in other organisms (Fig. 2A and C). Therefore, the known functions of the UBR box family may reveal new pathways in which the N-end rule pathway participates. As a founding UBR family member, UBR1 is solely responsible for the S. cerevisiae N-end rule pathway (6). S. cerevisiae UBR1 appears to have been duplicated during evolution into UBR1 and UBR2 in mammals but not in the fly or plants (see Fig. 3A). UBR1 and UBR2 have similar sizes (200 kDa) and sequences (46% identity), and N-terminal recognition specificities (45-47), and we anticipate that UBR1 and UBR2 cooperate in at least some biological processes. Consistent with this expectation is the previous finding that UBR1 and UBR2 form a stable complex with RECQL4 belonging to the RecQ helicase family that is implicated in the Rothmund-Thomson and Rapadilino syndromes (88).
We also recently found that UBR1 and UBR2 cooperate for in vivo degradation of RGS4 and RGS5, negative regulators of the G protein-coupled receptor (GPCR) signaling pathway (M. J. Lee, T. Tasaki, J. Y. An, K. Moroi, I. Davydov, and Y. T. Kwon, submitted for publication). Genetic and biochemical studies have implicated UBR1 and/or UBR2 in fat metabolism, male meiosis, female-specific development (in mice), peptide transport, and chromosome stability (in S. cerevisiae) (46, 47, 62, 74). UBR3 is a previously uncharacterized protein, and UBR3−/− neonatal offspring die in association with defects in milk suckling (T. Tasaki, A. Varshavsky, and Y. T. Kwon, unpublished data). Notably, we found that UBR3 is prominently expressed in the olfactory cortex of the brain, suggesting that UBR3 may be critical for the olfactory GPCR signaling pathway. Further characterization of UBR3−/− mice is under way. S. cerevisiae UBR2 (termed UBR3 in this study since it is a much stronger sequelog of mammalian UBR3 than UBR1/UBR2) was recently found to mediate the degradation of Rpn4, the transcriptional factor that regulates proteasome genes (81).
UBR4 has an extraordinary size (570 kDa) and appears to be a structural and functional homolog of Arabidopsis BIG (560 kDa) and Drosophila PUSHOVER (560 kDa) (19, 24, 37, 52, 54, 64, 87) (Fig. 3B). In contrast to other UBR box proteins, UBR4 lacks a known ubiquitylation domain. It is to be determined whether UBR4 is itself an E3 or a substrate recognition subunit of an unknown E3 complex. Arabidopsis BIG/a-UBR4 has been identified as two independent gene mutations which resulted in altered responses of hormone and light and severely impaired auxin transport (24, 37), suggesting its essential role in regulation of the function and transport of auxin, a major plant hormone (19, 24, 37, 52, 54). Drosophila PUSHOVER/UBR4 was originally identified as a calmodulin-binding protein (86). Genetic studies implicated PUSHOVER in male reproduction (64), nonrecombinant chromosome segregation in the female meiosis (67), synaptic transmission in photoreceptor cells (86), and perineurial glial growth by mediating signals from amn-encoded neuropeptide (87). Taken together, UBR4 and its homologs appear to regulate the signaling pathways in response to extrinsic stimuli such as hormones or neurotransmitters.
The 300-kDa UBR5 protein is a previously characterized mammalian E3 known as EDD (E3 identified by differential display) or hHYD (11, 29, 31, 65). UBR5 is a sequelog of the Drosophila tumor suppressor gene hyperplastic disks (55). UBR5 ubiquitylates TopBP1 for proteolysis (31) and interacts with CIB (calcium- and integrin-binding protein) in a DNA damage-dependent manner (29). TopBP1 is a member of the newly emerging BRCT (BRCA1 carboxyl-terminal) domain proteins implicated in DNA damage checkpoint and/or DNA repair pathways (90). CIB interacts with DNA-dependent protein kinase which functions in the DNA damage pathway (84).
UBR5 is often amplified in cancer and is implicated in cancer progression (16). More recently, knockout mice lacking EDD/UBR5 were found to be impaired in yolk sac vascularization (65). Drosophila HYD/d-UBR5 is required for fertility and functions as a tumor suppressor by controlling cell proliferation (55). Through experiments with a Drosophila cell line and the X-peptide pulldown assay, we demonstrated that, as would be expected from the findings with mouse UBR5, the insect UBR5 (HYD) protein is also, operationally, an N-recognin (see Results). Taken together, UBR5 and its homologs appear to be involved in DNA repair, cancer progression, and angiogenesis.
UBR6 is a 90-kDa orphan F-box protein with no apparent overall homology to other mouse proteins. The F-box protein is a component of the SCF E3 Ub ligase complex and contains two domains: an F-box motif that binds Skp1 for assembly into the SKP1/CUL1 complex and a carboxy-terminal domain that recognizes substrates (4, 82). The UBR box of UBR6 thus appears to be a substrate recognition domain for a putative SCFUBR6-E3 complex. UBR6 also contains the CASH domain, either a predicted or observed right-handed β-helix structure that is shared by many carbohydrate-binding proteins and sugar hydrolases (15). A splice valiant of human UBR6 was implicated in the human genetic disease called vitiligo (50).
UBR7 is a 50-kDa novel protein containing the PHD motif, a RING finger-like domain (26, 53). Various PHD proteins are known to be involved in chromatin-mediated transcriptional regulation (26). Overexpression of MLO2, an apparent UBR7 homolog in Schizosaccharomyces pombe, showed an asymmetrical segregation of chromosomes (34).
In summary, our identification of the UBR box E3 family unveils an enormous range of processes that are potentially mediated by the N-end rule pathway. It is to be investigated which of these functions are mediated by N-degron-dependent proteolysis.
Based on our findings, we propose a model in which the UBR box motif binds to N-degrons and other N-degron-like small molecules (e.g., amino acid derivatives, neurotransmitters, or hormones) to alter the protein ubiquitylation activity of the UBR box-containing E3 protein. Consistent with this model, a missense mutation of Arabidopsis BIG and UBR4 that perturbs auxin transport and growth was mapped to one of the conserved Cys residues within the UBR box motif (24) (Fig. 3A, shaded orange). Further, genetic analyses identified several S. cerevisiae UBR1 residues that are required for degradation of N-recognin substrates, and some of these critical residues (Cys145, Val146, Gly173, and Asp176 of S. cerevisiae UBR1) were a part of the UBR box motif (Fig. 3A, shaded pink) (A. Webster, M. Ghislain, and A. Varshavsky, unpublished data).
If the above conjecture proves correct, the known substrate-binding properties of UBR box proteins, including their being N-recognins, are likely to be a subset of their specificity repertoires. S. cerevisiae UBR1 contains three substrate-binding sites, two of which, type 1 and type 2, recognize (bind to) destabilizing N-terminal residues in proteins or short peptides (6, 22, 44, 74, 78). These sites target specific proteins bearing N-degrons for ubiquitylation, such as the separase-produced fragment of SCC1, a cohesin subunit (62). Remarkably, these two sites also function as nutritional/homeostatic sensors, through the same ability to bind polypeptides (including short peptides) with destabilizing N-terminal residues. This binding allosterically activates the third substrate-binding site of yeast UBR1, which recognizes an internal (non-N-terminal) degron of CUP9, a conditionally short-lived transcriptional repressor. CUP9 down-regulates several genes, including PTR2, which encodes a peptide transporter (10, 22, 74). The resulting positive-feedback circuit, in which imported peptides can activate the UBR1-dependent degradation of a repressor that down-regulates their import, allows S. cerevisiae to sense the presence of extracellular peptides and to react by accelerating their uptake (22, 74).
The conjectured diversity of recognition specificities of UBR-box E3s, if coupled with their differential, spatiotemporally controlled expression patterns, may underlie the already large and still expanding set of functions of the N-end rule pathway and other Ub-dependent pathways that involve UBR-box proteins.
Acknowledgments
We are grateful to Jai Wha Seo for establishment and immortalization of UBR1−/− UBR2−/− embryonic fibroblasts, Allen Shearn for anti-HYD antibody, and Song Li and Yong Wan for helpful advice.
This work was in part supported by NIH grant GM69482 and the American Heart Association Award (to Y.T.K), by NIH grants GM31530 and DK39520 and a grant from the Ellison Medical Foundation (to A. Varshavsky), by NIH grant AI47054 (to M.M.), and by NIH grant AI056987 to (L.C.F.M.).
REFERENCES
- 1.Aravind, L., L. M. Iyer, and E. V. Koonin. 2003. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle 2:123-126. [DOI] [PubMed] [Google Scholar]
- 2.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179-186. [DOI] [PubMed] [Google Scholar]
- 3.Bachmair, A., and A. Varshavsky. 1989. The degradation signal in a short-lived protein. Cell 56:1019-1032. [DOI] [PubMed] [Google Scholar]
- 4.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 5.Baker, R. T., and A. Varshavsky. 1995. Yeast N-terminal amidase. A new enzyme and component of the N-end rule pathway. J. Biol. Chem. 270:12065-12074. [DOI] [PubMed] [Google Scholar]
- 6.Bartel, B., I. Wunning, and A. Varshavsky. 1990. The recognition component of the N-end rule pathway. EMBO J. 9:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumeister, W., J. Walz, F. Zühl, and E. Seemüller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 8.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Woodbury, N.Y.
- 9.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 10.Byrd, C., G. C. Turner, and A. Varshavsky. 1998. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callaghan, M. J., A. J. Russell, E. Woollatt, G. R. Sutherland, R. L. Sutherland, and C. K. Watts. 1998. Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17:3479-3491. [DOI] [PubMed] [Google Scholar]
- 12.Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardozo, T., and M. Pagano. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell. Biol. 5:739-751. [DOI] [PubMed] [Google Scholar]
- 14.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 15.Ciccarelli, F. D., R. R. Copley, T. Doerks, R. B. Russell, and P. Bork. 2002. CASH-a beta-helix domain widespread among carbohydrate-binding proteins. Trends Biochem. Sci. 27:59-62. [DOI] [PubMed] [Google Scholar]
- 16.Clancy, J. L., M. J. Henderson, A. J. Russell, D. W. Anderson, R. J. Bova, I. G. Campbell, D. Y. Choong, G. A. Macdonald, G. J. Mann, T. Nolan, G. Brady, O. I. Olopade, E. Woollatt, M. J. Davies, D. Segara, N. F. Hacker, S. M. Henshall, R. L. Sutherland, and C. K. Watts. 2003. EDD, the human orthologue of the hyperplastic discs tumour suppressor gene, is amplified and overexpressed in cancer. Oncogene 22:5070-5081. [DOI] [PubMed] [Google Scholar]
- 17.Davydov, I. V., and A. Varshavsky. 2000. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem. 275:22931-22941. [DOI] [PubMed] [Google Scholar]
- 18.de Groot, R. J., T. Rümenapf, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 88:8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desgagné-Penix, I., S. Eakanunkul, J. P. Coles, A. L. Phillips, P. Hedden, and V. M. Sponsel. 2005. The auxin transport inhibitor response 3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. Plant J. 41:231-242. [DOI] [PubMed] [Google Scholar]
- 20.Ditzel, M., R. Wilson, T. Tenev, A. Zachariou, A. Paul, E. Deas, and P. Meier. 2003. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 5:467-473. [DOI] [PubMed] [Google Scholar]
- 21.Dohmen, R. J. 2000. Primary destruction signals, p. 188-205. In W. Hilt and D. Wolf (ed.), Proteasomes: the world of regulatory proteolysis. R. G. Landes Biosciences, Georgetown, Tex.
- 22.Du, F., F. Navarro-Garcia, Z. Xia, T. Tasaki, and A. Varshavsky. 2002. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA 99:14110-14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang, S., and A. M. Weissman. 2004. A field guide to ubiquitylation. Cell. Mol. Life. Sci. 61:1546-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gil, P., E. Dewey, J. Friml, Y. Zhao, K. C. Snowden, J. Putterill, K. Palme, M. Estelle, and J. Chory. 2001. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15:1985-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonda, D. K., A. Bachmair, I. Wünning, J. W. Tobias, W. S. Lane, and A. Varshavsky. 1989. Universality and structure of the N-end rule. J. Biol. Chem. 264:16700-16712. [PubMed] [Google Scholar]
- 26.Gozani, O., P. Karuman, D. R. Jones, D. Ivanov, J. Cha, A. A. Lugovskoy, C. L. Baird, H. Zhu, S. J. Field, S. L. Lessnick, J. Villasenor, B. Mehrotra, J. Chen, V. R. Rao, J. S. Brugge, C. G. Ferguson, B. Payrastre, D. G. Myszka, L. C. Cantley, G. Wagner, N. Divecha, G. D. Prestwich, and J. Yuan. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114:99-111. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton, M. H., L. A. Cook, T. R. McRackan, K. L. Schey, and J. D. Hildebrandt. 2003. γ2 subunit of G protein heterotrimer is an N-end rule ubiquitylation substrate. Proc. Natl. Acad. Sci. USA 100:5081-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heller, H., and A. Hershko. 1990. A ubiquitin-protein ligase specific for type III protein substrates. J. Biol. Chem. 265:6532-6535. [PubMed] [Google Scholar]
- 29.Henderson, M. J., A. J. Russell, S. Hird, M. Muñoz, J. L. Clancy, G. M. Lehrbach, S. T. Calanni, D. A. Jans, R. L. Sutherland, and C. K. Watts. 2002. EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J. Biol. Chem. 277:26468-26478. [DOI] [PubMed] [Google Scholar]
- 30.Hershko, A., A. Ciechanover, and A. Varshavsky. 2000. Basic Medical Research Award. The ubiquitin system. Nat. Med. 6:1073-1081. [DOI] [PubMed] [Google Scholar]
- 31.Honda, Y., M. Tojo, K. Matsuzaki, T. Anan, M. Matsumoto, M. Ando, H. Saya, and M. Nakao. 2002. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem. 277:3599-3605. [DOI] [PubMed] [Google Scholar]
- 32.Hondermarck, H., J. Sy, R. A. Bradshaw, and S. M. Arfin. 1992. Dipeptide inhibitors of ubiquitin-mediated protein turnover prevent growth factor-induced neurite outgrowth in rat pheochromocytoma PC12 cells. Biochem. Biophys. Res. Commun. 189:280-288. [DOI] [PubMed] [Google Scholar]
- 33.Iwamatsu, A. 1992. S-Carboxymethylation of proteins transferred onto polyvinylidene difluoride membranes followed by in situ protease digestion and amino acid microsequencing. Electrophoresis 13:142-147. [DOI] [PubMed] [Google Scholar]
- 34.Javerzat, J. P., G. Cranston, and R. C. Allshire. 1996. Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 24:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen, O. N., A. Podtelejnikov, and M. Mann. 1996. Delayed extraction improves specificity in database searches by matrix-assisted laser desorption/ionization peptide maps. Rapid Commun. Mass Spectrom. 10:1371-1378. [DOI] [PubMed] [Google Scholar]
- 36.Jin, J., T. Cardozo, R. C. Lovering, S. J. Elledge, M. Pagano, and J. W. Harper. 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanyuka, K., U. Praekelt, K. A. Franklin, O. E. Billingham, R. Hooley, G. C. Whitelam, and K. J. Halliday. 2003. Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J. 35:57-70. [DOI] [PubMed] [Google Scholar]
- 38.Kitamura, K., S. Katayama, S. Dhut, M. Sato, Y. Watanabe, M. Yamamoto, and T. Toda. 2001. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 1:389-399. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 40.Kwak, K. S., X. Zhou, V. Solomon, V. E. Baracos, J. Davis, A. W. Bannon, W. J. Boyle, D. L. Lacey, and H. Q. Han. 2004. Regulation of protein catabolism by muscle-specific and cytokine-inducible ubiquitin ligase E3α-II during cancer cachexia. Cancer Res. 64:8193-8198. [DOI] [PubMed] [Google Scholar]
- 41.Kwon, Y. T., S. A. Balogh, I. V. Davydov, A. S. Kashina, J. K. Yoon, Y. Xie, A. Gaur, L. Hyde, V. H. Denenberg, and A. Varshavsky. 2000. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol. Cell. Biol. 20:4135-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon, Y. T., A. S. Kashina, I. V. Davydov, R. G. Hu, J. Y. An, J. W. Seo, F. Du, and A. Varshavsky. 2002. An essential role of N-terminal arginylation in cardiovascular development. Science 297:96-99. [DOI] [PubMed] [Google Scholar]
- 43.Kwon, Y. T., A. S. Kashina, and A. Varshavsky. 1999. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol. Cell. Biol. 19:182-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon, Y. T., F. Lévy, and A. Varshavsky. 1999. Bivalent inhibitor of the N-end rule pathway. J. Biol. Chem. 274:18135-18139. [DOI] [PubMed] [Google Scholar]
- 45.Kwon, Y. T., Y. Reiss, V. A. Fried, A. Hershko, J. K. Yoon, D. K. Gonda, P. Sangan, N. G. Copeland, N. A. Jenkins, and A. Varshavsky. 1998. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 95:7898-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon, Y. T., Z. Xia, J. Y. An, T. Tasaki, I. V. Davydov, J. W. Seo, J. Sheng, Y. Xie, and A. Varshavsky. 2003. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 23:8255-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon, Y. T., Z. Xia, I. V. Davydov, S. H. Lecker, and A. Varshavsky. 2001. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol. Cell. Biol. 21:8007-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 49.Lawson, T. G., D. L. Gronros, P. E. Evans, M. C. Bastien, K. M. Michalewich, J. K. Clark, J. H. Edmonds, K. H. Graber, J. A. Werner, B. A. Lurvey, and J. M. Cate. 1999. Identification and characterization of a protein destruction signal in the encephalomyocarditis virus 3C protease. J. Biol. Chem. 274:9871-9880. [PubMed] [Google Scholar]
- 50.Le Poole, I. C., R. Sarangarajan, Y. Zhao, L. S. Stennett, T. L. Brown, P. Sheth, T. Miki, and R. E. Boissy. 2001. 'VIT1′, a novel gene associated with vitiligo. Pigment Cell Res. 14:475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lévy, F., N. Johnsson, T. Rümenapf, and A. Varshavsky. 1996. Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. USA 93:4907-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopéz-Bucio, J., E. Hernández-Abreu, L. Sánchez-Calderón, A. Pérez-Torres, R. A. Rampey, B. Bartel, and L. Herrera-Estrella. 2005. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 137:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9:945-956. [DOI] [PubMed] [Google Scholar]
- 54.Luschnig, C. 2001. Auxin transport: why plants like to think BIG. Curr. Biol. 11:R831-R833. [DOI] [PubMed] [Google Scholar]
- 55.Mansfield, E., E. Hersperger, J. Biggs, and A. Shearn. 1994. Genetic and molecular analysis of hyperplastic discs, a gene whose product is required for regulation of cell proliferation in Drosophila melanogaster imaginal discs and germ cells. Dev. Biol. 165:507-526. [DOI] [PubMed] [Google Scholar]
- 56.Mulder, L. C. F., and M. A. Muesing. 2000. Degradation of HIV-1 integrase by the N-end rule pathway. J. Biol. Chem. 275:29749-29753. [DOI] [PubMed] [Google Scholar]
- 57.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 58.Obin, M., E. Mesco, X. Gong, A. L. Haas, J. Joseph, and A. Taylor. 1999. Neurite outgrowth in PC12 cells. Distinguishing the roles of ubiquitylation and ubiquitin-dependent proteolysis. J. Biol. Chem. 274:11789-11795. [DOI] [PubMed] [Google Scholar]
- 59.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 60.Pickart, C. 2004. Back to the future with ubiquitin. Cell 116:181-190. [DOI] [PubMed] [Google Scholar]
- 61.Pickart, C., and D. Fushman. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8:610-616. [DOI] [PubMed] [Google Scholar]
- 62.Rao, H., F. Uhlmann, K. Nasmyth, and A. Varshavsky. 2001. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410:955-959. [DOI] [PubMed] [Google Scholar]
- 63.Rechsteiner, M., and C. P. Hill. 2005. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 15:27-33. [DOI] [PubMed] [Google Scholar]
- 64.Richards, S., T. Hillman, and M. Stern. 1996. Mutations in the Drosophila pushover gene confer increased neuronal excitability and spontaneous synaptic vesicle fusion. Genetics 142:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saunders, D. N., S. L. Hird, S. L. Withington, S. L. Dunwoodie, M. J. Henderson, C. Biben, R. L. Sutherland, C. J. Ormandy, and C. K. Watts. 2004. Edd, the murine hyperplastic disc gene, is essential for yolk sac vascularization and chorioallantoic fusion. Mol. Cell. Biol. 24:7225-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheel, H., and K. Hofmann. 2003. No evidence for PHD fingers as ubiquitin ligases. Trends Cell Biol. 13:285-287; author reply 287-288. [DOI] [PubMed] [Google Scholar]
- 67.Sekelsky, J. J., K. S. McKim, L. Messina, R. L. French, W. D. Hurley, T. Arbel, G. M. Chin, B. Deneen, S. J. Force, K. L. Hari, J. K. Jang, A. C. Laurencon, L. D. Madden, H. J. Matthies, D. B. Milliken, S. L. Page, A. D. Ring, S. M. Wayson, C. C. Zimmerman, and R. S. Hawley. 1999. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152:529-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sijts, A. J., I. Pilip, and E. G. Pamer. 1997. The Listeria monocytogenes-secreted p60 protein is an N-end rule substrate in the cytosol of infected cells. Implications for major histocompatibility complex class I antigen processing of bacterial proteins. J. Biol. Chem. 272:19261-19268. [DOI] [PubMed] [Google Scholar]
- 69.Solomon, V., S. H. Lecker, and A. L. Goldberg. 1998. The N-end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J. Biol. Chem. 273:25216-25222. [DOI] [PubMed] [Google Scholar]
- 70.Stary, S., X. J. Yin, T. Potuschak, P. Schlögelhofer, V. Nizhynska, and A. Bachmair. 2003. PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 133:1360-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki, T., and A. Varshavsky. 1999. Degradation signals in the lysine-asparagine sequence space. EMBO J. 18:6017-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taban, C. H., H. Hondermarck, R. A. Bradshaw, and B. Biolly. 1996. Effect of a dipeptide inhibiting ubiquitin-mediated protein degradation nerve-dependent limb regeneration in the newt. Experientia 52:865-870. [DOI] [PubMed] [Google Scholar]
- 73.Tobias, J. W., T. E. Shrader, G. Rocap, and A. Varshavsky. 1991. The N-end rule in bacteria. Science 254:1374-1377. [DOI] [PubMed] [Google Scholar]
- 74.Turner, G. C., F. Du, and A. Varshavsky. 2000. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405:579-583. [DOI] [PubMed] [Google Scholar]
- 75.Turner, G. C., and A. Varshavsky. 2000. Detecting and measuring cotranslational protein degradation in vivo. Science 289:2117-2120. [DOI] [PubMed] [Google Scholar]
- 76.Ui-Tei, K., Y. Naito, F. Takahashi, T. Haraguchi, H. Ohki-Hamazaki, A. Juni, R. Ueda, and K. Saigo. 2004. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32:936-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varshavsky, A. 2003. The N-end rule and regulation of apoptosis. Nat. Cell Biol. 5:373-376. [DOI] [PubMed] [Google Scholar]
- 78.Varshavsky, A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93:12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varshavsky, A. 2004. ‘Spalog’ and ‘sequelog’: neutral terms for spatial and sequence similarity. Curr. Biol. 14:R181-R183. [DOI] [PubMed] [Google Scholar]
- 80.Varshavsky, A. 2000. Ubiquitin fusion technique and its descendants. Methods Enzymol. 327:578-593. [DOI] [PubMed] [Google Scholar]
- 81.Wang, L., X. Mao, D. Ju, and Y. Xie. 2004. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 279:55218-55223. [DOI] [PubMed] [Google Scholar]
- 82.Winston, J. T., D. M. Koepp, C. Zhu, S. J. Elledge, and J. W. Harper. 1999. A family of mammalian F-box proteins. Curr. Biol. 9:1180-1182. [DOI] [PubMed] [Google Scholar]
- 83.Wolf, D. H., and W. Hilt. 2004. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim. Biophys. Acta 1695:19-31. [DOI] [PubMed] [Google Scholar]
- 84.Wu, X., and M. R. Lieber. 1997. Interaction between DNA-dependent protein kinase and a novel protein, KIP. Mutat. Res. 385:13-20. [DOI] [PubMed] [Google Scholar]
- 85.Xie, Y., and A. Varshavsky. 1999. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 18:6832-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu, X. Z., P. D. Wes, H. Chen, H. S. Li, M. Yu, S. Morgan, Y. Liu, and C. Montell. 1998. Retinal targets for calmodulin include proteins implicated in synaptic transmission. J. Biol. Chem. 273:31297-31307. [DOI] [PubMed] [Google Scholar]
- 87.Yager, J., S. Richards, D. S. Hekmat-Scafe, D. D. Hurd, V. Sundaresan, D. R. Caprette, W. M. Saxton, J. R. Carlson, and M. Stern. 2001. Control of Drosophila perineurial glial growth by interacting neurotransmitter-mediated signaling pathways. Proc. Natl. Acad. Sci. USA 98:10445-10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yin, J., Y. T. Kwon, A. Varshavsky, and W. Wang. 2004. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 13:2421-2430. [DOI] [PubMed] [Google Scholar]
- 89.Yoshida, S., M. Ito, J. Gallis, I. Nishida, and A. Watanabe. 2002. A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyl-transferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 32:129-137. [DOI] [PubMed] [Google Scholar]
- 90.Yu, X., C. C. Chini, M. He, G. Mer, and J. Chen. 2003. The BRCT domain is a phospho-protein binding domain. Science 302:639-642. [DOI] [PubMed] [Google Scholar]
- 91.Yuan, B., R. Latek, M. Hossbach, T. Tuschl, and F. Lewitter. 2004. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32:W130-W134. [DOI] [PMC free article] [PubMed] [Google Scholar]