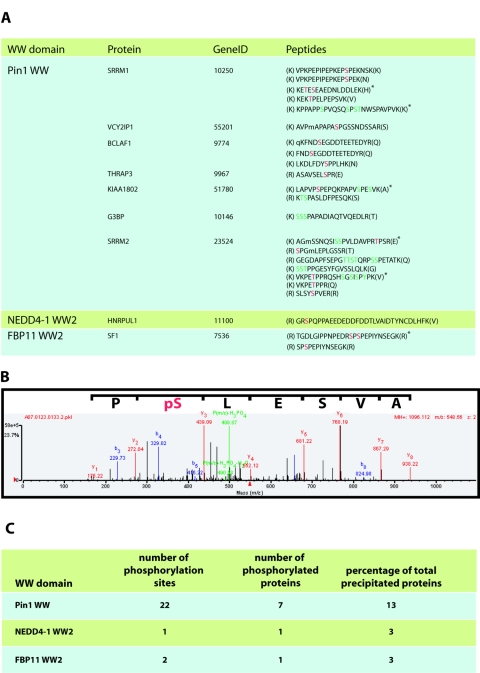
FIG. 5.
Analysis of GST-WW domain pulldowns for phosphopeptides. (A) The MS data from the GST-Pin1 WW, NEDD4-1 WW2, and FBP11 WW2 pulldowns were analyzed for the presence of phosphopeptides as described in Materials and Methods. Identified phosphopeptides and their host proteins are indicated. Definitively phosphorylated residues are indicated in red, and possible phosphorylation sites where the identification was ambiguous are indicated in green. A lowercase “m” denotes an oxidized methionine, while a lowercase “q” denotes a pyroglutamine residue. Amino acids in parentheses are residues N and C terminal to the peptide in the protein sequence. Peptides indicated by an asterisk are doubly phosphorylated. (B) The MS/MS spectra for the THRAP3 phosphopeptide is shown. Note that only b (blue), y (red), and parental ion minus phosphoric acid (P(m/z) − H3PO4) or minus phosphoric acid and water (P(m/z) − H3PO4-H2O) (green) are indicated. The number associated with each ion is the mass/charge ratio (m/z) of that ion. (C) Statistics showing the number of phosphorylation sites, number of phosphorylated proteins, and number of phosphorylated proteins as a percentage of total proteins precipitated with each WW domain (from Fig. 3).