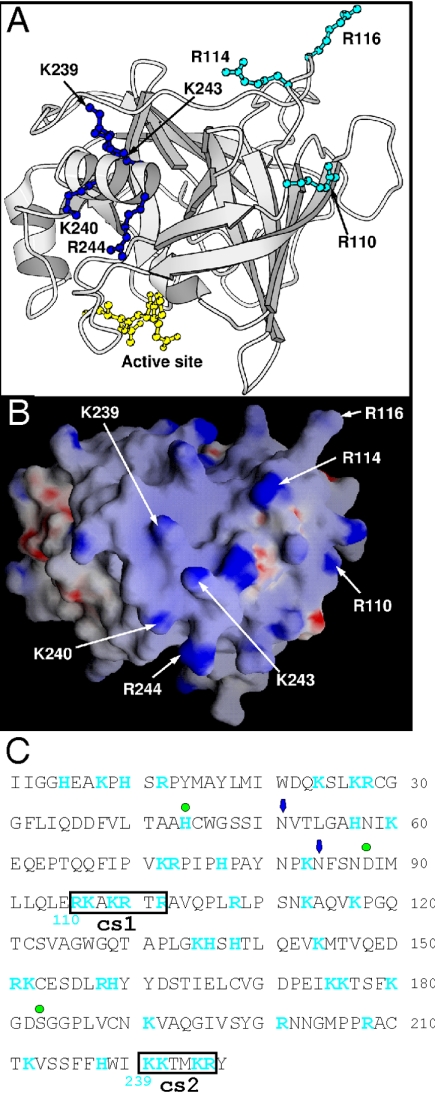
FIG. 3.
Location of two clusters of positively charged residues on human GrB. The structure of human GrB bound to a peptide inhibitor has been solved (39) (Protein Database accession number: 1IAU). (A) The residues mutated to alanine in the GrB cs mutants are labeled (numbering in 1IAU is based by convention on chymotrypsin). R110, R114, and R116 (cs1) are in cyan ball-and-stick, and K239, K240, K243, and K244 (cs2) are in blue ball-and-stick. The peptide inhibitor Ac-IEPD-CHO is in yellow ball-and-stick, and indicates the position of the catalytic site. (B) GRASP electrostatic potential surface of GrB in an orientation similar to the image shown in panel A. The positively charged residues selected for mutagenesis are labeled. (C) Sequence of mature GrB showing positively charged residues (blue) and the cationic sequences (boxed). Green dots mark the residues of the catalytic triad, and arrows mark the positions of Asn-linked carbohydrate side chains. Numbering in blue below the sequence is based on chymotrypsin.