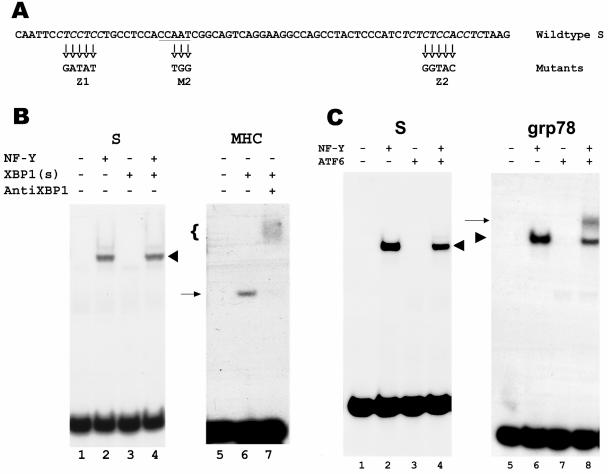
FIG. 5.
Lack of binding of XBP1(s) and ATF6-alpha to the HBV S promoter. A. Sequence of the S promoter, with the CCAAT element (underlined) and the CT-rich elements (italics) (52). The mutated sequences in the M2 and Z1+Z2 mutants are shown below. B. EMSA of the S promoter, using in vitro-synthesized XBP1(s) in the absence or presence of NF-Y (lanes 3 and 4, respectively). The shifted band resulting from binding by NF-Y is marked with a chevron. As a negative control, luciferase synthesized in vitro from the plasmid luciferase T7 control DNA (Promega) was substituted for XBP1(s) (lanes 1 and 2). As a positive control, the MHC DRA promoter X box was used to bind XBP1(s) (lane 6). The shifted band is marked with an arrow, and the presence of XBP1(s) in this band was confirmed by supershifting with antibodies to XBP1 (lane 7, marked with a bracket). C. EMSA of the S promoter, using in vitro-synthesized ATF6-alpha in the absence or presence of NF-Y (lanes 3 and 4, respectively). The shifted band resulting from binding by NF-Y is marked with a chevron. As a negative control, in vitro-synthesized luciferase was substituted for ATF6-alpha (lanes 1 and 2). As a positive control, the grp78 promoter ERSE was used to bind NF-Y alone, ATF6-alpha alone, or both (lanes 6, 7, and 8, respectively). The band resulting from binding by NF-Y alone is marked with a chevron, and the band resulting from binding by both NF-Y and ATF6-alpha is marked with an arrow.