Abstract
In self-renewing tissues such as the skin epidermis and the bone marrow, Myc proteins control differentiation of stem cells and proliferation of progenitor cell types. In the epithelium of the small intestine, we show that c-Myc and N-Myc are expressed in a differential manner. Whereas c-Myc is expressed in the proliferating transient-amplifying compartment of the crypts, N-Myc is restricted to the differentiated villus epithelium and a single cell located near the crypt base. c-Myc has been implicated as a critical target of the canonical Wnt pathway, which is essential for formation and maintenance of the intestinal mucosa. To genetically assess the role of c-Myc during development and homeostasis of the mammalian intestine we induced deletion of the c-mycflox allele in the villi and intestinal stem cell-bearing crypts of juvenile and adult mice, via tamoxifen-induced activation of the CreERT2 recombinase, driven by the villin promoter. Absence of c-Myc activity in the juvenile mucosa at the onset of crypt morphogenesis leads to a failure to form normal numbers of crypts in the small intestine. However, all mice recover from this insult to form and maintain a normal epithelium in the absence of c-Myc activity and without apparent compensation by N-Myc or L-Myc. This study provides genetic and molecular evidence that proliferation and expansion of progenitors necessary to maintain the adult intestinal epithelium can unexpectedly occur in a Myc-independent manner.
Precise control of cell renewal, lineage commitment, differentiation, and apoptosis are all required to maintain the homeostasis of the mammalian intestinal tract. In the mouse, the basic structure of the intestinal epithelium is established during late gestation (9, 29, 51). Around embryonic day (E) 16, the upward growth of underlying mesenchymal tissue creates the nascent, fingerlike epithelial villi that protrude into the lumen of the gut. This formation is concomitant with physical separation of the differentiated epithelial cells to the nascent villi and proliferative epithelial cells to the lower regions that separate respective villi. Intervillus regions are the precursor structures of the crypts of Lieberkühn that form around day 7 of postnatal life (P7) by invagination through the mesenchyme towards the lining of the gut. These structures (crypts and intervillus regions) also contain the self-renewing intestinal stem cells (ISCs) that permanently produce a population of rapidly proliferating progenitor cells that migrate toward the lumen of the intestine, where they undergo cell cycle arrest and commit to different cell lineages by terminal differentiation (31, 44).
The differentiation of progenitor cells in the intestine is restricted to four different cell fates-the absorptive enterocytes and the secretory goblet, enteroendocrine, and Paneth cell lineages. The migration of differentiated intestinal cells follows a bidirectional pathway, where goblet cells, enteroendocrine cells, and enterocytes migrate from the intervillus regions or crypt towards the villi, while Paneth cells descend to the base of the crypts. Differentiation of proliferative intestinal cells is accompanied by down-regulation of c-myc expression and up-regulation of the cell cycle inhibitor p21cip/waf, which is concomitant with cell cycle arrest (38, 53). Differentiated cells in the villi perform their respective functions and are then sloughed into the lumen, following induction of apoptosis, after some 2 to 5 days (6, 8, 10, 17, 42, 45).
These events entail the normal developmental and homeostatic events of the mammalian intestine. Recent reports suggest that intestinal development and homeostasis are largely controlled via the canonical Wnt pathway. Interestingly, the proto-oncogene c-myc has been proposed as a key downstream Wnt target gene that may control proliferation, differentiation, and transformation in the adult intestine (26, 38, 53). However, the endogenous role of c-Myc in the adult intestine is unknown.
The c-myc gene encodes a short-lived transcription factor (c-Myc) with a basic/helix-loop-helix/leucine zipper (bHLH-LZ) domain, that mediates DNA binding and heterodimerization with its partner Max. c-Myc/Max heterodimers activate or repress two distinct pools of target genes that elicit a variety of biological responses, including cell cycle progression, cellular growth, differentiation, and apoptosis/survival (2, 12, 18, 28, 32, 37). Deletion of c-myc by gene targeting in mice causes midgestation lethality, and mutant embryos fail to develop a primitive hematopoietic system (11, 32, 52). Therefore, assessment of the role of c-Myc in specific cell and tissue types in the adult or juvenile mouse requires a conditional knockout approach.
Recently, we have shown that conditional loss of c-myc in the adult bone marrow severely disrupts normal homeostasis of the hematopoietic system. Hematopoietic stem cells lacking a functional copy of c-myc self-renew but fail to initiate normal differentiation, resulting in the accumulation of hematopoietic stem cells in situ combined with severe cytopenia (56). Conversely, overexpression of c-myc in skin or hematopoietic stem cells leads to loss of the stem cell pool, presumably via premature differentiation (55, 56). These reports highlight a central role for c-Myc in the balance between stem cell self-renewal, differentiation, and maintenance of homeostasis in at least some self-renewable tissues (32). However, despite the known expression of c-Myc in the proliferating cells of the intestinal crypts, the role of c-Myc during the development and homeostasis of this tissue remains unknown.
MATERIALS AND METHODS
Mice.
c-mycΔORF/+ mice (52) were crossed with transgenic villin::CreERT2 mice (13), to generate villin::CreERT2, c-mycΔORF/+ founders. These mice were crossed with c-mycflox/flox mice (52) to generate experimental (villin::CreERT2, c-mycΔORF/flox) and control (c-mycΔORF/flox, c-myc+/flox or villin::CreERT2; c-mycΔORF/+) mice. Mice were injected intraperitoneally with 1 mg of tamoxifen per 20 g body weight into 7-day postnatal mice for 4 consecutive days. We assessed the efficiency of the deletion of c-myc by Taqman PCR of cDNA and genomic DNA from scrapings of intestinal tissue. Mice were of mixed genetic background and only littermates were used for the analysis.
RNA analysis.
Deletion of the c-myc allele was assessed by Taqman real-time PCR on scrapings of intestinal tissue that separate the crypt and villi from surrounding tissue. Taqman reverse transcription (RT)-PCR was also performed to analyze c-myc, L-myc and N-myc expression. Primers and conditions were as described (52), except for L-myc, where primers LmycF (ACGGCACTCGTCTGGAA) and LmycR (GTGACTGGCTTTCGGATGTC) were used. All cDNAs were normalized using β2microglobulin expression with β2mF (GTGTATGCTATCCAGAAAACCC) and β2mR (TCACATGTCTCGATCCCAGTAG).
Southern blotting.
DNA was isolated from epithelial scrapings of intestinal tissue, digested with HindIII and XbaI and hybridized with 32P-labeled probes as described (52). The c-myc probe was generated by PCR amplification from pATmyc11 using cmycsouthp 1(ATTCCAGCGAGAGACAGAGG) and cmycsouthp 2 (CCAACATCAAGTCCATGTGC).
Histology, immunohistochemistry, and bromodeoxyuridine labeling.
All tissues were fixed in 10% formalin or 4% paraformaldehyde, paraffin embedded, and sectioned at 3 to 6 μm for hematoxylin/eosin staining or immunostaining procedures as previously described (3). The primary antibodies used were rabbit anti-FabpL (1:500; kind gift of Jeffery Gordon), rabbit antisynptophysin (Dako; 1:100), rat anti-CD44v6 (1:100; Pharmingen), rabbit antilysozyme (1:500; DAKO), mouse anti-Ki67 (1:100; Novocastra), mouse antibromodeoxyuridine (1:500; Sigma), mouse anti-p21cip/waf (1:100, Santa Cruz), rabbit anti-c-Myc (1:500; Upstate Biotechnology) or anti-c-Myc (1:250; N262, SantaCruz), and rabbit anti-N-Myc (1:200; Santa Cruz). The peroxidase-conjugated secondary antibodies used were mouse or rabbit EnVision+ (Dako) or mouse anti-rat-horseradish peroxidase (1:250; Biosource). For bromodeoxyuridine labeling, mice were injected with 100 μg of bromodeoxyuridine (Sigma) per gram of body weight and sacrificed 2 h postinjection.
RESULTS
Expression of c-Myc and N-Myc in the intestinal epithelium.
Previous studies have detected c-myc and N-myc expression in E14.5 gut epithelium. At E16.5 of development c-myc transcripts are restricted to the proliferative cells of the embryonic gut while N-myc transcripts localize to the differentiated cell types of the villi (22). Expression of both c-myc and N-myc, but not L-myc, is detectable by Northern blotting in the newborn and adult intestine (47, 57). We have further investigated expression of Myc family members in the intestine using real-time PCR on whole pieces of proximal duodenal tissue derived from wild-type 8-day-old (P8) mice or epithelial cell scrapings from P14 and P60 (adult) mice. Similar to what has been reported previously (38, 47, 53, 57), we detected c-myc and N-myc expression in the duodenum of the small intestine in all age groups of mice, while L-myc expression was not detected above background levels (data not shown).
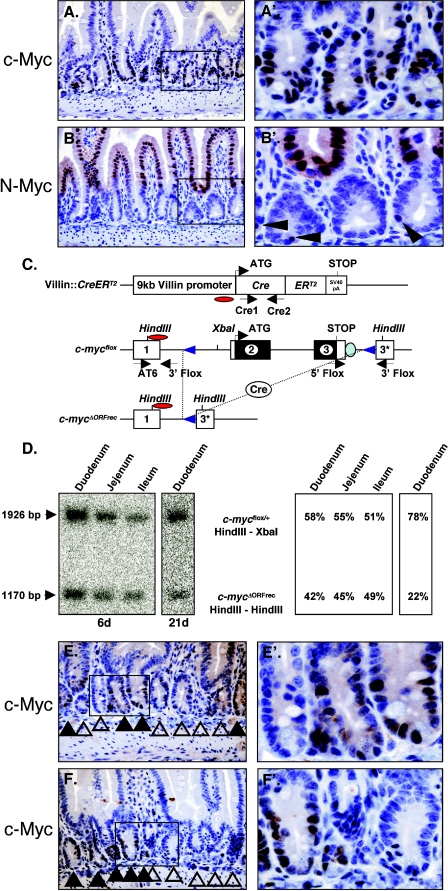
To determine the specific expression pattern of c-Myc and N-Myc proteins in the intestinal epithelium, immunohistochemical analysis was performed. This analysis revealed c-Myc expression in the proliferating cells of the juvenile and adult crypts, confirming the results of others (Fig. 1A) (38, 53). In contrast, juvenile mice expressed high N-Myc in differentiated epithelial cells of the villi, which gradually decreased as mice matured (Fig. 1B and data not shown). In addition, individual N-Myc-expressing cells located at the bottom of the crypts directly adjacent to Paneth cells were detected in some crypts (Fig. 1B). Taken together these data show that with the exception of the Paneth cells, all cells of the crypt/villus axis express either c-Myc or N-Myc. Their expression appears mutually exclusive and the c-Myc expression domain of the crypt is flanked by differentiated N-Myc-expressing epithelial cells.
FIG.1.
Myc expression and villin::CreERT2-mediated recombination of the c-mycflox allele in the small intestine. (A) Immunohistochemical analysis of c-Myc in the adult duodenum. Enlargement of the indicated box is shown in A′. (B and B′) Immunohistochemistry of N-Myc in adult duodenum. (C) Schematic representation of the villin::CreERT2 and c-mycflox alleles used. The c-mycΔORFrec allele represents the locus after Cre mediated recombination. The three c-myc exons are illustrated as boxes; regions in black indicate the open reading frame. Restriction sites are shown on top. Red ovals indicate probe used for Southern blot analysis. Arrows below each scheme indicate locations of the primers used for genotyping. (D) Southern blot analysis of villin::CreERT2; c-mycflox/+ mice in the small intestine 6 days (duodenum, ileum, and jejunum) and 21 days (duodenum) after the first of four consecutive tamoxifen injections. The c-myc probe hybridizes to the floxed and wild-type alleles (1,926 bp) and to the deleted c-mycΔORFrec allele (ΔORFrec: 1,170 bp) in recombined cells. (E and F) Immunohistochemical analysis of c-Myc in duodenal tissue sections derived from villin::CreERT2, c-mycΔORF/flox mice 21 days following the first injection of tamoxifen. Solid arrows indicate c-Myc-expressing crypts (c-mycΔORF/flox). Open arrows indicate c-Myc-negative crypts (c-mycΔORF/ORFrec).
Conditional ablation of c-myc in the crypts of the mouse small intestine.
To genetically investigate the role of c-Myc during small intestinal development and maintenance, we used mice carrying the c-mycflox allele (Fig. 1C), which has its open reading frame flanked by loxP sites (52). To induce c-Myc elimination in the small intestine, villin::CreERT2 mice were used, in which the villin promoter targets the tamoxifen-inducible CreERT2 recombinase to the intestinal epithelium (13). To first assess the recombination efficiency of villin::CreERT2 at the c-mycflox locus, villin::CreERT2; mycflox/+ mice were generated. Instead of using mice with two c-mycflox alleles, animals with only a single copy of the c-mycflox allele were chosen to avoid putative selection against cells losing all c-Myc activity. After tamoxifen-induced Cre activity in P7 pups recombination was assessed by Southern blotting using DNA obtained from intestinal epithelial scrapings (containing epithelium and villin-negative mesenchyme) 6, 21, and 90 days post-first tamoxifen injection (dpi).
The c-myc probe utilized hybridizes to the floxed and wild-type alleles (1,926 bp) and to the deleted c-mycΔORFrec allele (ΔORFrec: 1,170 bp) in recombined cells (Fig. 1C and D). Thus, a 50% loss of the c-mycflox allele in c-mycflox/+ mice is equivalent to 100% recombination. At 6 dpi mutant mice showed 84% (2 × 42%) recombination in the duodenum and a 90% and 98% recombination in the jejunum and ileum, respectively (Fig. 1D). These data indicate that shortly after the termination of tamoxifen injection, villin::CreERT2 has induced high-level recombination of the c-mycflox allele in the small intestine. This is expected due to the strong expression of villin in the differentiated epithelium of the villus as well as crypt cells (13, 39). However, by 21 dpi, the recombination level in duodenum drops to 44% (Fig. 1D) and remains at similar levels thereafter (data not shown). Shortly after the completion of tamoxifen injections CreERT2 becomes inactive as tamoxifen is degraded and over time only cells derived from either recombined or unrecombined ISCs will populate the intestine. The persistence of recombined cells over time demonstrates recombination of the c-mycflox allele in close to half of all crypt based ISCs (13). This explains the drop in recombination frequency that is observed shortly after the completion of tamoxifen injections.
To further determine the extent of c-Myc protein elimination immunohistochemical analysis was performed on villin::CreERT2; c-mycflox/ΔORF mice following tamoxifen injection (mutant mice). As shown in Fig. 1E and F, a significant number of crypts in mutant mice lack c-Myc expression even three weeks after Cre induction. Crypts in mutant mice contained either many c-Myc-expressing cells or none at all. Intervillus regions of juvenile mice are initially polyclonal with respect to ISCs but by 2 weeks of age a crypt progenitor cell and its descendants displace all other cells from the crypt to attain stem cell based monoclonality (40, 46). This monoclonality of crypts allows visual distinction between ISCs, suggesting clonal derivation from either c-Myc+ (escapers) or c-Myc− (deleted) ISCs. Since both types of crypts appear indistinguishable c-Myc is dispensable for normal intestinal epithelium homeostasis (Fig. 1E and F and 2A and B).
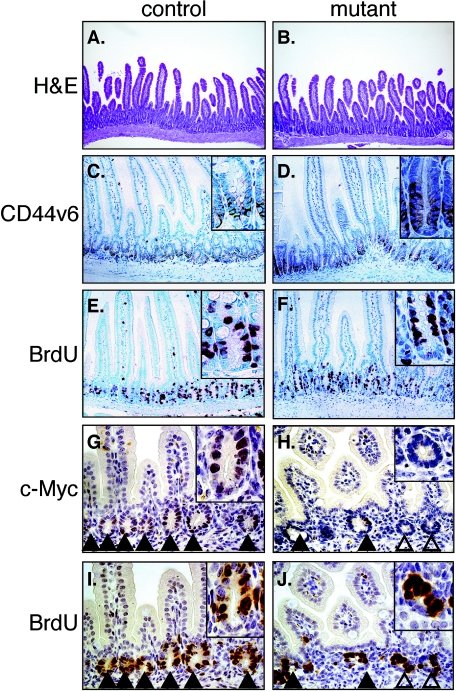
FIG. 2.
Histological analysis and marker gene expression in small intestines of control (A, C, E, G, and I) and mutant (B, D, F, H, and J) mice. (A and B) Hematoxylin/eosin staining. Magnification, 100×. (C and D) Immunohistochemical analysis of CD44v6 expression. (E and F) Bromodeoxyuridine (BrdU) incorporation. (G to J) Immunohistochemical analysis of serial sections of the duodenum detecting c-Myc expression (G and H) and bromodeoxyuridine incorporation (I and J). Solid arrows indicate c-Myc-expressing crypts (c-mycΔORFrec/flox) and open ones indicate c-Myc-negative crypts (c-mycΔORFrec/ΔORFrec). (A to F) Adult mice (G to J) P14 juvenile mice. Magnification, 200×.
Induced loss of c-Myc activity in small intestinal crypts does not alter cell proliferation or fate of crypt/villus cells.
The differentiation fate of ISCs in the small intestine is limited to the formation of either adsorptive enterocytes or the secretory cell lineages of goblet cells, enteroendocrine cells, or crypt-based Paneth cells (50). Recently, it has been shown that inhibition of in vivo Wnt signaling in the small intestine, through the use of transgenic mice ectopically expressing the secreted Wnt inhibitor Dickkopf1 (Dkk1) inhibits the formation of secretory cell lineages without affecting the formation of enterocytes. Furthermore, inhibition of Wnt signaling leads to a reduction of epithelial proliferation and a greatly reduced number of visible crypts. Disrupted intestinal homeostasis was accompanied by strongly reduced c-myc expression, reduced cellular proliferation and a subsequent up-regulation of the cell cycle inhibitor and c-Myc target p21cip1/waf1 (38).
To assess the effect of c-Myc loss on the fate and proliferation of intestinal crypt cells, small intestines derived from mice injected with tamoxifen at P7 were assessed for crypt formation and expression of numerous markers. Adult mice showed no change in small intestinal development between control and mutant mice (Fig. 2A and B). Sections from mice 21 dpi were assessed for expression of the crypt marker CD44v6 and incorporation of bromodeoxyuridine. All crypts in mutant duodenum retain normal expression of the crypt marker CD44v6 (Fig. 2C and D) and proliferate normally as assessed by bromodeoxyuridine incorporation (Fig. 2E and F). Unfortunately, the antigen retrieval systems differ between c-Myc and the other markers, preventing double labeling with c-Myc. Despite the fact that c-Myc negative crypts were present in all mutant mice as was regularly assessed by immunohistochemistry (Fig. 1E and Fig. 5A and B and data not shown), serial sections were performed to explicitly demonstrate that proliferation was unaffected in the crypts lacking c-Myc expression (Fig. 2G to J).
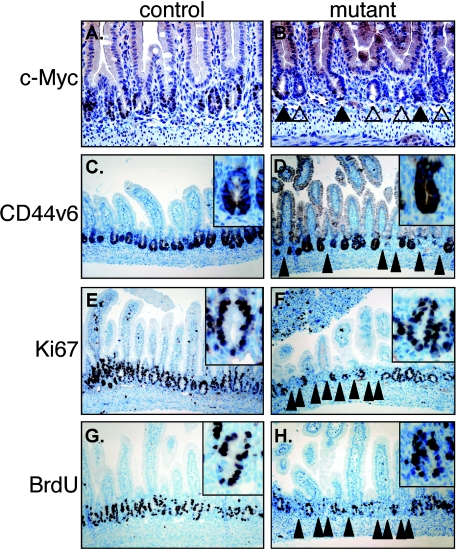
FIG. 5.
Immunohistochemical analysis of c-Myc and crypt markers in juvenile duodenal tissue sections of control (A, C, E, and G) and mutant (B, D, F, and H) mice 6 days after Cre induction. (A and B) c-Myc; (C and D) CD44v6; (E and F) Ki67; (G and H) bromodeoxyuridine (BrdU). Solid arrows indicate c-Myc-positive crypts. Open arrows indicate c-Myc negative crypts. Solid pointed arrowheads in panels D, F, and H indicate areas lacking normal crypt formation.
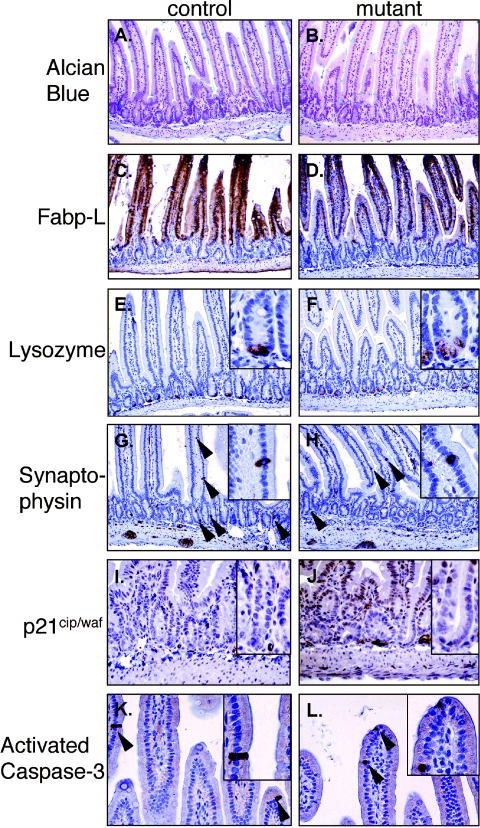
To identify the differentiation status of small intestine mosaic for c-myc expression, sections derived from control and mutant mice 21 dpi, were stained for the goblet cell markers Pas/Schiff (data not shown) and Alcian blue (Fig. 3A and B) or subjected to immunohistochemical analysis for the enterocyte marker fatty acid-binding protein liver (Fabp-L) (Fig. 3C and D) (49) the Paneth cell marker lysozyme (Fig. 3E and F) and the enteroendocrine cell marker synaptophysin (Fig. 3G and H). Immunohistochemical analysis of these markers showed that both control and mutant small intestine have normal production of both enteroyctes and secretory lineages. In addition, loss of c-myc did not alter the expression of the cell cycle inhibitor p21Cip1/Waf1 (Fig. 3I and J). Collectively, these data indicate that loss of c-Myc does not alter differentiation or cell fate in the small intestine.
FIG. 3.
Expression analysis of differentiation and apoptosis markers in control (A, C, E, G, I, and K) and mutant (B, D, F, H, J, and L) duodenum. (A and B) Alcian Blue staining to detect goblet cells. (C and D) Fabp-L expression to detect enterocytes. (E and F) Lysozyme expression in Paneth cells. (G and H) Synaptophysin expression in endocrine cells. (I and H) Expression of the CDK inhibitor p21cip/waf. (K and L) expression of activated caspase-3 to monitor apoptotic cells. Arrows indicate lysozyme, synaptophysin or activated caspase-3-positive epithelial cells.
Apoptosis is a fundamental cellular process for the homeostasis of self-renewable tissues and overexpression of c-myc in normal cells is a potent inducer of apoptosis while fibroblasts lacking c-Myc are resistant to certain apoptotic stimuli (1, 4, 14, 15). In the intestine differentiated cells apoptose and slough into the lumen as they reach the tip of the villi. To assess the apoptotic pathway in mammalian intestine lacking c-Myc, we performed immunohistochemical analysis for the cleaved form of caspase-3 as a marker of cellular apoptosis (35) on duodenal tissue derived from mice 6, 10, 14, 21 and 90 days after c-Myc deletion (day 21 results shown in Fig. 3K and L). Activated caspase-3 positive cells were seen at the tip, and/or middle sections, of a small and equivalent number of villi in both control and mutant mice. These data indicate that loss of c-Myc in the small intestine does not alter the normal apoptotic fate of differentiated cells.
c-Myc is required for the induction of crypt formation.
During the first two weeks of life, the postnatal intervillus regions invaginate to form the crypts. These intervillus regions contain the actively dividing, c-Myc-expressing TA cells and ISCs. Crypt formation commences around day 7 and culminates around the suckling/weaning transition, when the crypts acquire their mature structure (21 days postnatal) (10). Little is known of the process of intestinal crypt invagination. In the skin, invagination of hair follicles requires the action of the canonical Wnt pathway acting through alterations in cadherin gene expression (24). c-myc is a known target of the Wnt pathway in intestinal cells (19, 38, 53) and has been shown to alter the expression of cell adhesion genes such as cadherins and integrins in the skin and bone marrow (16, 55, 56).
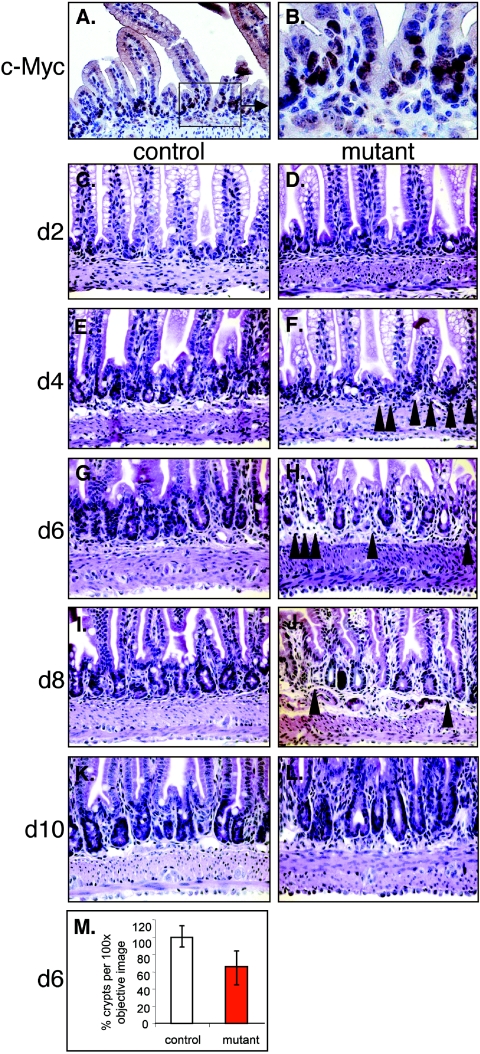
Immunohistochemical analysis of histological tissue sections showed expression of c-Myc at P8 (Fig. 4A and B). To investigate the role of c-Myc during crypt invagination in normal and mutant duodenum, pups were injected with tamoxifen at P7, sacrificed and intestines histologically analyzed 2, 4, 6, 8 and 10 dpi. At 2 dpi, no disorganization of intervillus regions was observed (Fig. 4C and D) while mice sacrificed between 4 and 8 dpi showed numerous regions with a reduced number of invaginating crypts when compared to control intestines (Fig. 4E to H and M and 5C to H and Table 1). Quantification of the number of crypts in histological sections at 6 dpi revealed a 34.2 ± 18.7% (n = 395/600) decrease in crypt number in mutant duodenum across 6 littermate pairs of control and mutant mice (Fig. 4M). Unexpectedly, by 10 to 90 dpi both mutant and control mice show indistinguishable intestinal crypt morphology (Fig. 4L). Table 1 shows an analysis on the prevalence of the crypt formation defect over time.
FIG. 4.
Kinetic analysis of the crypt formation process in normal (A, B, C, E, G, I, and K) and mutant (D, F, H, J, and L) mice. (A and B) c-Myc expression in the intervillus region of seven-day-old mice. (C to L) Seven-day-old control or mutant mice were injected with tamoxifen and the duodenum was analyzed by hematoxylin/eosin staining every second day for 10 days. (M) Graphical representation of the percentage of crypts per phase contrast image (100× objective) of tissue sections derived from six different littermates of control (100%) and mutant mice (red bar) 6 days after the first tamoxifen injection. Solid arrows in F, H, and J indicate areas in which crypt formation has not occurred.
TABLE 1.
Prevalence of the reduced crypt number phenotype
| Day after first tamoxifen injection | Age of mice (days) | No. of mutant mice with reduced crypt numbers/no. examined |
|---|---|---|
| 2 | 9 | 0/5 |
| 4 | 11 | 3/4 |
| 6 | 13 | 8/9 |
| 8 | 15 | 4/7 |
| 10 | 17 | 0/2 |
| 14 | 21 | 0/2 |
| 21 | 28 | 0/2 |
| 90 | 97 | 0/3 |
Highly proliferative epithelial cells are involved in the crypt invagination process. Since c-Myc is thought to be a master regulator of cell division, the proliferative status of these cells was investigated in normal and mutant duodenum. Immunohistochemical analysis for c-Myc expression showed that mutant mice either expressed c-Myc in most or in none of their TA cells, suggesting a clonal origin of the mutant crypts or a clonal expression pattern of villinCreERT2 (Fig. 5A and B). Both c-Myc positive and c-Myc negative crypts expressed the crypt marker CD44v6 (Fig. 5C and D), the proliferation markers Ki67 (Fig. 5. E,F) and incorporated bromodeoxyuridine (Fig. 5G and H and Fig. 2G to J). While this marker analysis underscores again the failure to form crypts in numerous areas of the intestine (arrows in Fig. 5D, F, and H), all crypts that do form in mutants (carrying a mix between c-Myc-expressing and -nonexpressing crypts) display a consistent number of Ki67 or bromodeoxyuridine positive cells per crypt (control: 10.0 ± 2; mutant: 9.2 ± 1.1). These data indicate that deletion of c-Myc activity, just prior to crypt invagination, inhibits normal crypt formation but does not affect proliferation and maintenance of already formed crypts.
Expression of N-myc in the intestine following loss of c-Myc expression.
The transient nature of the crypt formation phenotype as well as the absence of long-term defects in mouse intestine lacking a functional copy of the c-myc gene suggests redundancy by other Myc family members. N-myc has been shown to functionally replace c-myc in vivo (30) suggesting that the presence of N-Myc in crypt cells lacking c-Myc could provide a functional rescue. However, N-Myc is normally not present in the crypt cell types expressing c-Myc (Fig. 1A). RNA analysis using real-time RT-PCR did not show any significant increase of N-Myc in c-Myc mutant intestinal epithelia isolated from 2, 6, and 21 dpi (data not shown).
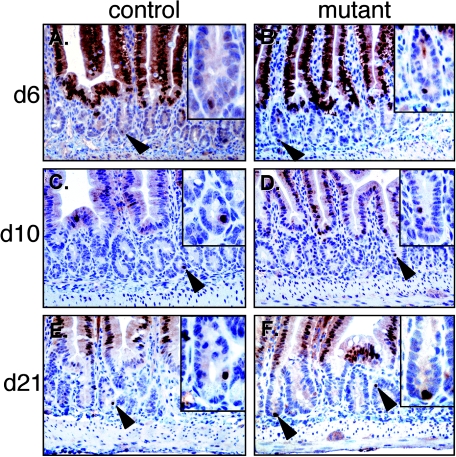
To further investigate whether N-Myc protein is ectopically expressed in c-Myc-deficient crypts immunohistochemical analysis on sections from c-Myc mutants (6, 10, and 21 dpi) was performed. As shown in Fig. 6, N-myc expression in mutant intestine remained restricted to the villi and a single cell at the bottom of the crypt. Importantly, no significant ectopic N-Myc expression was detected in the c-Myc expression domain. In addition L-myc transcripts did not increase above background levels detected in normal intestinal tissue (data not shown) (47). Taken together these data suggest that proliferation and maintenance of the intestinal crypts are not dependent on Myc (c-Myc, L-Myc, and N-Myc) activity.
FIG. 6.
Immunohistochemical analysis of N-Myc expression in control (A, C, and E) or mutant (B, D, and F) duodenum after Cre induction at the time points indicated. Arrowheads indicate single N-Myc-expressing cells at the bottom of crypts.
DISCUSSION
With the exception of Paneth cells, all epithelial cells of the crypt-villus unit express either N-Myc or c-Myc. The strong expression of c-Myc in the proliferative cells of the crypt and its known role in proliferation and differentiation has strongly suggested a significant role for this factor during intestinal homeostasis. However, unexpectedly this study demonstrates that conditional loss of functional c-myc in the intestinal mucosa of adult and juvenile mice has no effect on the normal homeostasis of this rapidly self-renewing tissue. These results are surprising in light of a number of studies, suggesting c-Myc is a key regulator of crypt proliferation and differentiation. For example, expression profiling of microdissected early progenitor populations from intestinal crypts had identified a number of genes either controlling or controlled by c-Myc (50).
In addition, c-myc has also been identified as a target of the canonical Wnt-β-catenin-TCF4 pathway (19, 53). Inhibition of this pathway in the mouse intestinal mucosa, via overexpression of the extracellular Wnt inhibitor Dkk1, revealed a phenotype in adult mice with a reduced number of crypts, concomitant with a loss of proliferation (27, 38). Loss of proliferating cells correlated with loss of c-Myc expression and an increase in expression of p21cip/waf, a well-known target gene repressed by c-Myc. Finally, mice lacking the Wnt signaling component TCF4 show a proliferation defect of perinatal crypt stem/progenitors concomitant with a lack of c-Myc expression (26, 53). Collectively, these data indicated that the Wnt pathway plays a role in the regulation of crypt homeostasis and cellular proliferation in the intestine and that c-myc is a likely candidate target gene that controls these processes.
Tight control of Wnt pathway signaling and c-Myc expression levels has also been demonstrated to alter the differentiation fate of intestinal cells. Mouse intestine inhibited in Wnt pathway signaling fails to form secretory lineages correctly (26, 38), while in vitro cultures of colorectal carcinoma cells that respond to Wnt signaling can be induced to have a twofold difference of c-myc levels that determines whether these cells proliferate or differentiate (53). In contrast, results presented here show that loss of c-Myc in intestinal crypt cells in vivo does not lead to an alteration of cell fate away from secretory cell lineages and that expression of p21cip/waf, epithelial cell differentiation, and apoptosis remain unchanged. This suggests that other TCF4 target genes must mediate the essential function of the canonical Wnt signaling pathway during intestinal development and maintenance (53).
The lack of a notable homeostatic phenotype in the small intestine of c-myc-deficient mice could most easily be explained by a compensatory up-regulation of another Myc family member. N-Myc has been previously shown to functionally replace c-Myc in vivo (30) and in contrast to L-Myc, N-Myc is expressed in the intestine (Fig. 1B) making this protein a likely candidate. However, in the normal intestinal epithelium N-Myc expression is spatially distinct from the c-Myc expression domain and is restricted to the differentiated cells of the intestinal villi and a single cell at the crypt base. These data indicate that N-Myc and c-Myc in the intestinal epithelium seem to be expressed in a differential and possibly mutually exclusive manner. In addition, despite the fact that c-myc;N-myc double-knockout intestines need to be generated to formally exclude the possibility of N-Myc compensation for c-Myc deficiency, neither N-Myc nor L-Myc is ectopically expressed in the c-Myc expression domain of mutant intestines, making it very unlikely that the lack of a phenotype is due to a compensatory mechanism. Instead, our data indicate the presence of a crypt proliferation program independent of general Myc activity.
Similarly, hematopoietic stem cells in the bone marrow proliferate without c-Myc, while more differentiated progenitors depend on c-Myc activity for cell cycle progression. Recent genetic data obtained from mouse mutants lacking several components of the cell cycle machinery such as cyclins D1 to D3, cyclins E1 and E2, or CDKs 4 and 6 also suggest a more complex cell cycle regulation in vivo compared to what is known from studies in cultured cells (48). It remains to be shown whether several independent cell cycle programs exist in vivo or whether the system is highly redundant and thus can compensate by various means for the lack of any active component, making the system highly flexible and extraordinarily stable.
Deletion of c-myc in juvenile mice leads to a temporary reduction in crypt numbers.
Specific deletion of c-myc in juvenile mice induced a temporary failure to form correct numbers of invaginated crypts within the small intestine. This phenotype was transient in nature and appeared in small but distinct regions of the intestine. Affected regions of tissue occurred with a high penetrance and were only observed if mice were injected around the commencement of crypt morphogenesis, which initiates around 7 days of age (54). All mice recover to form histologically normal intestines.
This is not the first report of transcription factor loss causing an effect on crypt morphogenesis. Homozygous loss of the Ets transcription factor Elf3 has been reported to have inhibitory effects on mouse crypt morphogenesis in the absence of effects on cellular proliferation or apoptosis. However, this phenotype is also coupled with abnormal extracellular matrix deposition and differentiation (34). While little is known about the morphogenesis of crypts, invagination of hair follicles in the skin is known to be regulated via the Wnt pathway and is dependent on a change in expression of E-cadherin to P-cadherin on the surface of invaginating cells. Forced elevation of E-cadherin levels in the skin blocks follicle invagination (24). However, this mechanism is unlikely to be in place during crypt morphogenesis, as forced expression of E-cadherin along the length of the crypt-villus axis in the mouse small intestinal epithelium does not alter differentiation or crypt morphogenesis but does slow cell migration and proliferation and induce apoptosis in the crypt (21). In this respect it is highly relevant to note that adhesion molecules and other proteins involved in migration and cytoskeletal organization are controlled by c-Myc (16, 56). Future studies, using less mosaic Cre lines, need to investigate whether some of these may be crucially involved in the failure of crypt invagination of c-Myc mutants.
c-Myc as a specific intestinal cancer target.
The c-myc gene is overexpressed in approximately one out of five human cancers (33). In addition, c-myc has been identified as a target of the canonical Wnt-β-catenin-TCF4 pathway (19), and 85% of human intestinal tumors contain activating mutations in this pathway (3). Interestingly, 70% of these intestinal tumors have up-regulated c-myc expression, without significant gene amplification (5). These data suggest that uncontrolled Wnt-induced activation of cell proliferation requires c-Myc activity. The direct role of c-Myc in the development of adenomatous polyposis coli (APC)-like tumors in mice is currently under investigation. However, our data showing that c-Myc is not essential for the homeostasis of the crypt and intestine would suggest that specific targeting of c-Myc function in the intestine of patients with APC-activated intestinal tumors may be deleterious to tumor proliferation without affecting normal intestinal homeostatic mechanisms.
Myc and intestinal stem cells.
Our recent report on the deletion of c-myc in the bone marrow revealed a requirement for this proto-oncogene in controlling the balance between hematopoietic stem cell self-renewal and differentiation by inhibition of differentiation while stem cell self-renewal remains c-Myc independent. This phenotype is mediated by interactions with the stem cell niche and is apparent despite the presence of detectable N-Myc expression, suggesting a specific role for c-Myc and potentially also N-Myc in adult stem cell biology. A role for c-Myc in the control of progenitor cell fate, independent of its known function in division and survival, has also been uncovered in other systems such as the neural crest and the skin epidermis (7, 32, 55).
Recent studies using microarray analysis of various stem cell populations revealed N-Myc as a general stem cell marker (23, 43). It is therefore intriguing that we have uncovered N-Myc expression in individual cells at a position previously suggested to be the location of ISCs (31). Very little is known about how ISCs are controlled. Conditional inactivation of the Bmpr1a gene in the mouse intestine has recently been shown to increase the number of putative ISCs, which in turn leads to an increased number of crypts and intestinal polyposis (20). Analysis of this phenotype uncovered a homeostatic mechanism in the intestine regulated by a complex interaction between the canonical BMP and Wnt pathways as well as activity of the tumor suppressor Pten. This study also suggested novel putative ISC markers including BmpR1a, 14-3-3ζ, P-Pten, and P-Akt, which, together with N-Myc, may complement previously suggested markers such as Musashi-1 and Hes1 (25, 36, 41). Thus, the elimination of N-Myc function in the intestinal epithelium will be crucial to shed more light on the role of the Myc family members in adult tissue stem cell biology in vivo.
Acknowledgments
We thank the laboratory of Hans Clevers for help setting up immunohistochemistry and Jeffrey Gordon for the Fabp-L antibody. We thank Sandra Offner for technical assistance and members of the Trumpp laboratory for helpful discussions and comments on the manuscript.
This work was supported by grants to A.T. from the Swiss National Science Foundation, the Swiss Cancer League, and the Leenards Foundation. A.T. is member of the EMBO Young Investigator Programme. M.B. was supported by a fellowship from the Roche Research Foundation.
REFERENCES
- 1.Alarcon, R. M., B. A. Rupnow, T. G. Graeber, S. J. Knox, and A. J. Giaccia. 1996. Modulation of c-Myc activity and apoptosis in vivo. Cancer Res. 56:4315-4319. [PubMed] [Google Scholar]
- 2.Amati, B., K. Alevizopoulos, and J. Vlach. 1998. Myc and the cell cycle. Front Biosci. 3:D250-68. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, C. B., K. L. Neufeld, and R. L. White. 2002. Subcellular distribution of Wnt pathway proteins in normal and neoplastic colon. Proc. Natl. Acad. Sci. USA 99:8683-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askew, D. S., R. A. Ashmun, B. C. Simmons, and J. L. Cleveland. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6:1915-1922. [PubMed] [Google Scholar]
- 5.Augenlicht, L. H., S. Wadler, G. Corner, C. Richards, L. Ryan, A. S. Multani, S. Pathak, A. Benson, D. Haller, and B. G. Heerdt. 1997. Low-level c-myc amplification in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 57:1769-1775. [PubMed] [Google Scholar]
- 6.Batlle, E., J. T. Henderson, H. Beghtel, M. M. van den Born, E. Sancho, G. Huls, J. Meeldijk, J. Robertson, M. van de Wetering, T. Pawson, and H. Clevers. 2002. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111:251-263. [DOI] [PubMed] [Google Scholar]
- 7.Bellmeyer, A., J. Krase, J. Lindgren, and C. LaBonne. 2003. The protooncogene c-Myc is an essential regulator of neural crest formation in Xenopus. Dev. Cell 4:827-839. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, H., and C. P. Leblond. 1974. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. III. Entero-endocrine cells. Am. J. Anat. 141:503-519. [DOI] [PubMed] [Google Scholar]
- 9.Colony, P. C., and M. R. Neutra. 1985. Macromolecular transport in the fetal rat intestine. Gastroenterology 89:294-306. [DOI] [PubMed] [Google Scholar]
- 10.Crossman, M. W., S. M. Hauft, and J. I. Gordon. 1994. The mouse ileal lipid-binding protein gene: a model for studying axial patterning during gut morphogenesis. J. Cell Biol. 126:1547-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, A. C., M. Wims, G. D. Spotts, S. R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671-682. [DOI] [PubMed] [Google Scholar]
- 12.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 13.el Marjou, F., K. P. Janssen, B. H. Chang, M. Li, V. Hindie, L. Chan, D. Louvard, P. Chambon, D. Metzger, and S. Robine. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39:186-193. [DOI] [PubMed] [Google Scholar]
- 14.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 15.Facchini, L. M., and L. Z. Penn. 1998. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 12:633-651. [PubMed] [Google Scholar]
- 16.Frye, M., C. Gardner, E. R. Li, I. Arnold, and F. M. Watt. 2003. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development 130:2793-2808. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, J. I., and M. L. Hermiston. 1994. Differentiation and self-renewal in the mouse gastrointestinal epithelium. Curr. Opin. Cell Biol. 6:795-803. [DOI] [PubMed] [Google Scholar]
- 18.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 19.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 20.He, X. C., J. Zhang, W. G. Tong, O. Tawfik, J. Ross, D. H. Scoville, Q. Tian, X. Zeng, X. He, L. M. Wiedemann, Y. Mishina, and L. Li. 2004. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 36:1117-1121. [DOI] [PubMed] [Google Scholar]
- 21.Hermiston, M. L., M. H. Wong, and J. I. Gordon. 1996. Forced expression of E-cadherin in the mouse intestinal epithelium slows cell migration and provides evidence for nonautonomous regulation of cell fate in a self-renewing system. Genes Dev. 10:985-996. [DOI] [PubMed] [Google Scholar]
- 22.Hirning, U., P. Schmid, W. A. Schulz, G. Rettenberger, and H. Hameister. 1991. A comparative analysis of N-myc and c-myc expression and cellular proliferation in mouse organogenesis. Mech. Dev. 33:119-125. [DOI] [PubMed] [Google Scholar]
- 23.Ivanova, N. B., J. T. Dimos, C. Schaniel, J. A. Hackney, K. A. Moore, and I. R. Lemischka. 2002. A stem cell molecular signature. Science 298:601-604. [DOI] [PubMed] [Google Scholar]
- 24.Jamora, C., R. DasGupta, P. Kocieniewski, and E. Fuchs. 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayahara, T., M. Sawada, S. Takaishi, H. Fukui, H. Seno, H. Fukuzawa, K. Suzuki, H. Hiai, R. Kageyama, H. Okano, and T. Chiba. 2003. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 535:131-135. [DOI] [PubMed] [Google Scholar]
- 26.Korinek, V., N. Barker, P. Moerer, E. van Donselaar, G. Huls, P. J. Peters, and H. Clevers. 1998. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19:379-383. [DOI] [PubMed] [Google Scholar]
- 27.Kuhnert, F., C. R. Davis, H. T. Wang, P. Chu, M. Lee, J. Yuan, R. Nusse, and C. J. Kuo. 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 101:266-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levens, D. L. 2003. Reconstructing MYC. Genes Dev. 17:1071-1077. [DOI] [PubMed] [Google Scholar]
- 29.Madara, J. L., M. R. Neutra, and J. S. Trier. 1981. Junctional complexes in fetal rat small intestine during morphogenesis. Dev. Biol. 86:170-178. [DOI] [PubMed] [Google Scholar]
- 30.Malynn, B. A., I. M. de Alboran, R. C. O'Hagan, R. Bronson, L. Davidson, R. A. DePinho, and F. W. Alt. 2000. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 14:1390-1399. [PMC free article] [PubMed] [Google Scholar]
- 31.Marshman, E., C. Booth, and C. S. Potten. 2002. The intestinal epithelial stem cell. Bioessays 24:91-98. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, M. J., A. Wilson, and A. Trumpp. 2005. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 3:128-137. [DOI] [PubMed]
- 33.Nesbit, C. E., J. M. Tersak, and E. V. Prochownik. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18:3004-3016. [DOI] [PubMed] [Google Scholar]
- 34.Ng, A. Y., P. Waring, S. Ristevski, C. Wang, T. Wilson, M. Pritchard, P. Hertzog, and I. Kola. 2002. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology 122:1455-1466. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson, D. W., A. Ali, N. A. Thornberry, J. P. Vaillancourt, C. K. Ding, M. Gallant, Y. Gareau, P. R. Griffin, M. Labelle, Y. A. Lazebnik, and et al. 1995. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376:37-43. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura, S., N. Wakabayashi, K. Toyoda, K. Kashima, and S. Mitsufuji. 2003. Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig. Dis. Sci. 48:1523-1529. [DOI] [PubMed] [Google Scholar]
- 37.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 38.Pinto, D., A. Gregorieff, H. Begthel, and H. Clevers. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto, D., S. Robine, F. Jaisser, F. E. El Marjou, and D. Louvard. 1999. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J. Biol. Chem. 274:6476-6482. [DOI] [PubMed] [Google Scholar]
- 40.Ponder, B. A., G. H. Schmidt, M. M. Wilkinson, M. J. Wood, M. Monk, and A. Reid. 1985. Derivation of mouse intestinal crypts from single progenitor cells. Nature 313:689-691. [DOI] [PubMed] [Google Scholar]
- 41.Potten, C. S., C. Booth, G. L. Tudor, D. Booth, G. Brady, P. Hurley, G. Ashton, R. Clarke, S. Sakakibara, and H. Okano. 2003. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation 71:28-41. [DOI] [PubMed] [Google Scholar]
- 42.Potten, C. S., and M. Loeffler. 1990. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development 110:1001-1020. [DOI] [PubMed] [Google Scholar]
- 43.Ramalho-Santos, M., S. Yoon, Y. Matsuzaki, R. C. Mulligan, and D. A. Melton. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298:597-600. [DOI] [PubMed] [Google Scholar]
- 44.Sancho, E., E. Batlle, and H. Clevers. 2003. Live and let die in the intestinal epithelium. Curr. Opin. Cell Biol. 15:763-770. [DOI] [PubMed] [Google Scholar]
- 45.Sancho, E., E. Battle, and H. Clevers. 2004. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20:695-723. [DOI] [PubMed]
- 46.Schmidt, G. H., D. J. Winton, and B. A. Ponder. 1988. Development of the pattern of cell renewal in the crypt-villus unit of chimaeric mouse small intestine. Development 103:785-790. [DOI] [PubMed] [Google Scholar]
- 47.Semsei, I., S. Y. Ma, and R. G. Cutler. 1989. Tissue and age specific expression of the myc proto-oncogene family throughout the life span of the C57BL/6J mouse strain. Oncogene 4:465-471. [PubMed] [Google Scholar]
- 48.Sherr, C. J., and J. M. Roberts. 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18:2699-2711. [DOI] [PubMed] [Google Scholar]
- 49.Simon, T. C., K. A. Roth, and J. I. Gordon. 1993. Use of transgenic mice to map cis-acting elements in the liver fatty acid-binding protein gene (Fabpl) that regulate its cell lineage-specific, differentiation-dependent, and spatial patterns of expression in the gut epithelium and in the liver acinus. J. Biol. Chem. 268:18345-18358. [PubMed] [Google Scholar]
- 50.Stappenbeck, T. S., J. C. Mills, and J. I. Gordon. 2003. Molecular features of adult mouse small intestinal epithelial progenitors. Proc. Natl. Acad. Sci. USA 100:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trier, J. S., and P. C. Moxey. 1979. Morphogenesis of the small intestine during fetal development. Ciba Found. Symp. 70:3-29. [DOI] [PubMed] [Google Scholar]
- 52.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 53.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 54.Vidrich, A., J. M. Buzan, C. Ilo, L. Bradley, K. Skaar, and S. M. Cohn. 2004. Fibroblast growth factor receptor-3 is expressed in undifferentiated intestinal epithelial cells during murine crypt morphogenesis. Dev. Dyn. 230:114-123. [DOI] [PubMed] [Google Scholar]
- 55.Waikel, R. L., Y. Kawachi, P. A. Waikel, X. J. Wang, and D. R. Roop. 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 28:165-168. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, A., M. J. Murphy, G. M. Oser, T. Oskarsson, K. Kaloulis, M. D. Bettess, A. C. Pasche, C. Knabenhans, H. R. MacDonald, and A. Trumpp. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmerman, K. A., G. D. Yancopoulos, R. G. Collum, R. K. Smith, N. E. Kohl, K. A. Denis, M. M. Nau, O. N. Witte, D. Toran-Allerand, C. E. Gee, et al. 1986. Differential expression of myc family genes during murine development. Nature 319:780-783. [DOI] [PubMed] [Google Scholar]