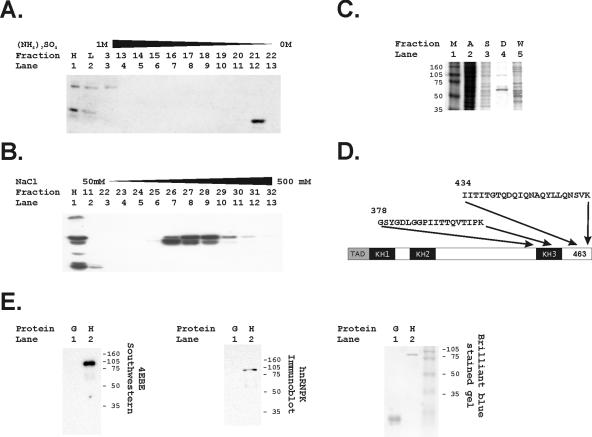
FIG. 3.
Purification of the 68-kDa protein binding to the 4EBE by DNA affinity chromatography identifies hnRNP K. (A) Hydrophobic exchange chromatography was performed as the initial step in protein purification. Fractions identifying the 68-kDa species binding to the 4EBE were identified by Southwestern analyses as described in Materials and Methods. Lane 1 contains a positive control HeLa nuclear extract (H), and lane 2 contains an aliquot of the HeLa extract (L) before loading on the column. (B) The second step in purification used anion exchange chromatography. Fractions identifying the 68-kDa species binding to the 4EBE were identified by Southwestern analyses as described in Materials and Methods. The positive control HeLa extract was again included for comparison (H). (C) Two sequential bindings to a DNA affinity chromatography matrix using a trimeric 4EBE sequence purified a 68-kDa band as demonstrated in this brilliant blue-stained acrylamide gel. Size markers (M, lane 1), an aliquot of the positive fraction from the hydrophobic exchange column (A, lane 2), an aliquot of the positive fraction from the monoS column (S, lane 3), and an aliquot of the DNA affinity wash (W, lane 5) are compared to the protein bound to the 4EBE-DNA affinity matrix (D) shown in lane 4. (D) The 68-kDa protein band was submitted for sequencing and the indicated amino acid sequences were identified. These correspond uniquely to hnRNP K. (E) Bacterially synthesized gst-hnRNP K was run in a standard Southwestern analysis, and the trimeric 4EBE probe bound to the gst-hnRNP K (H) but not to glutathione S-transferase alone (G). This band was confirmed to be hnRNP K by an immunoblot analysis with anti-hnRNP K. Finally, the gel was also stained with brilliant blue to demonstrate loading of the species tested for binding to the 4EBE.