Fig. 2. Schematic outlining of protein identification by mass spectrometric analysis.
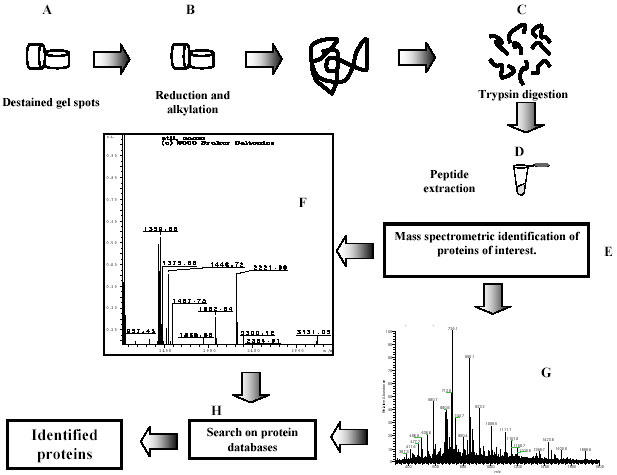
After 2-D PAGE, the protein spots were excised, subjected to distaining (A), and reduction and alkylation reactions (B), prior to digestion with trypsin (C). The volume was adjusted to ~20 μl (D), at this point the sample was ready for mass spectrometry analysis (E). An aliquot of the supernatant containing tryptic peptides is analyzed by MALDI-TOF MS, which results in a peptide-mass fingerprint of the protein; the panel shows the individual peptide peaks in the mass spectrum (F). The reminder of the supernatant is desalted and analyzed by nanoelectrospray ionization tandem-MS (ESI-MS). The figure shows how the sequence can be derived by fragmentation of the same peptide by tandem mass spectrometry (G).
