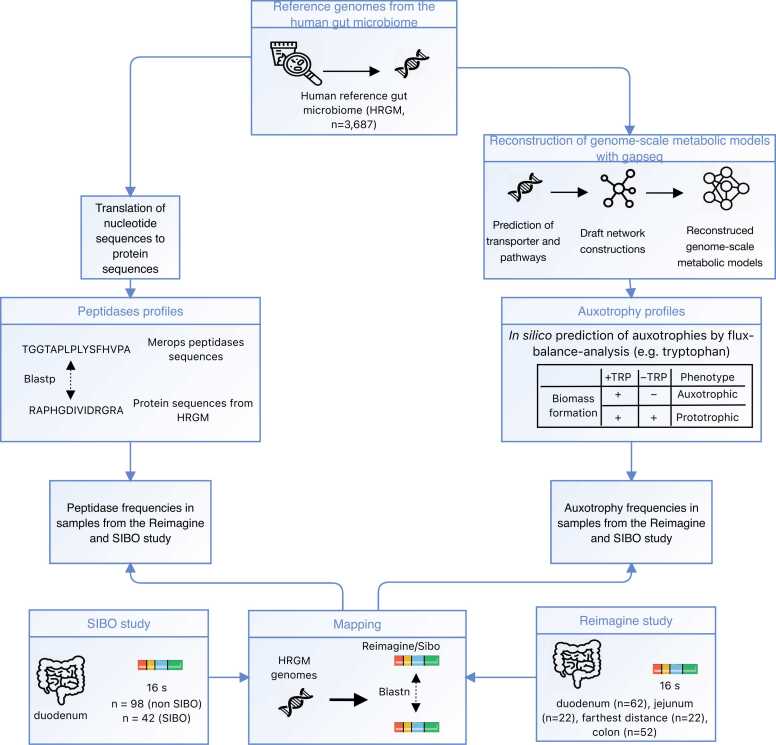
Fig. 1.
Flowchart of the study. Genome-scale metabolic models were reconstructed from genomes of the human reference gut microbiome [38] (HRGM) catalog with gapseq [39]. Auxotrophies were predicted with metabolic modeling using flux-balance analysis. The peptidase profiles were determined by scanning the peptidase sequences of the HRGM catalog for peptidases from the MEROPS database [40]. The distribution of peptidases and auxotrophies was predicted in the SIBO and Reimagine study cohorts after mapping on HRGM genomes. The farthest distance refers to the distance that could be reached during the sampling. Free available icons were taken from www.flaticon.com (creators: photo3idea_studio, surang, Icon home Eucalyp, Kiranshastry, Becris, Nadiinko).