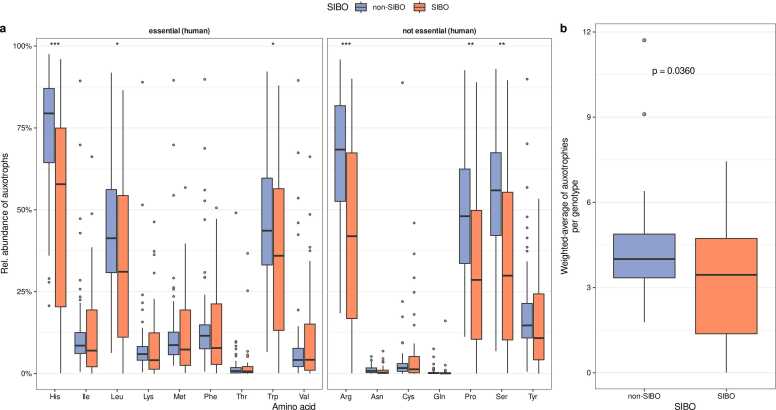
Fig. 5.
Genome-scale metabolic modeling predicted auxotrophy profiles in the duodenal microbiomes of non-SIBO and SIBO samples. (a) Individual amino acid auxotrophy abundances in the duodenal microbiome of SIBO and non-SIBO patients. Asterisks denote the statistical significance of differences in the relative abundance of the respective auxotrophy between the SIBO and the non-SIBO group (Wilcoxon rank sum test, * p < 0.05, ** p < 0.01, *** p < 0.001). p values are FDR-adjusted. (b) Abundance-weighted average of the number of auxotrophies per gut microbial genotype in SIBO and non-SIBO (Wilcoxon rank sum test, p = 0.036).