Abstract
The global production and use of antimicrobial chemicals surged during and after the COVID-19 pandemic, yet their developmental toxicity in aquatic organisms at environmentally relevant concentrations remains poorly understood. Here, we investigate and compare the developmental effects of two restricted antimicrobial chemicals—triclosan (TCS) and triclocarban (TCC)—and three alternative antimicrobials—benzalkonium chloride (BAC), benzethonium chloride (BEC), and chloroxylenol (CX)—on zebrafish embryos (Danio rerio) at concentrations of 0.4, 4, and 40 μg L−1. We find that BAC induces the most severe reduction in hatching rates, followed by TCS, TCC, BEC, and CX. BAC also exhibits the strongest inhibition of heart rate, with toxicity levels comparable to those of TCS and TCC. All tested chemicals, except CX, cause significant teratogenic effects. Transcriptomic analysis reveals substantial disruptions in immune-related coagulation cascades and mitogen-activated protein kinase signaling pathways. Further validation via protein-protein interaction network analysis and real-time quantitative polymerase chain reaction confirms that altered expression of key hub genes in these pathways impacts bone and heart development, as well as immune system function, potentially driving developmental toxicity. This study provides the first systematic comparison of developmental toxicity among currently used antimicrobials at environmentally relevant concentrations, revealing that the alternative antimicrobial BAC poses greater developmental risks than the banned TCS and TCC. These findings raise concerns about the safety of BAC as a widespread substitute and highlight the necessity for more rigorous environmental risk assessments of alternative antimicrobials before their large-scale application.
Keywords: Disinfectants, Hatchability, Developmental toxicity, Transcriptomic analysis, Zebrafish
Graphical abstract

Highlights
-
•
Antimicrobial chemicals cause teratogenic effects in zebrafish embryos.
-
•
The alternative antimicrobial BAC exhibits toxicity comparable to that of banned chemicals TCS and TCC.
-
•
Antimicrobial chemicals disrupt immune-related and MAPK signaling pathways.
1. Introduction
Antimicrobial chemicals have broad-spectrum activity against bacteria, fungi, and viruses; therefore, they are commonly used in home and personal care products and pharmaceuticals, as well as employed in the agricultural, food processing, and clinical health care settings, to control the spread of environmental pathogens [[1], [2], [3]]. For example, triclosan (TCS), triclocarban (TCC), benzalkonium chloride (BAC), benzethonium chloride (BEC), and chloroxylenol (CX) are antimicrobial chemicals commonly used in existing disinfection products [3]. Regulatory actions, such as the Food and Drug Administration's ban on TCS and TCC in antibacterial soaps, have prompted a shift to alternatives, such as BAC, BEC, and CX [3,4]. The coronavirus disease 2019 (COVID-19) pandemic markedly increased their use in disinfection practices across diverse settings, including hospitals, homes, and workplaces, to curb the transmission of severe acute respiratory syndrome coronavirus 2 [[5], [6], [7]]. The widespread production and use of these antimicrobial chemicals have resulted in their common detection in various environmental matrices, such as indoor residential dust, surface water, sewage sludge, sediments, and human tissue [[8], [9], [10], [11], [12], [13], [14], [15]]. The reported concentrations of TCS and TCC in surface water have reached up to 40 and 6.75 μg L−1, respectively [16,17]. Moreover, while data on BEC concentrations in environmental matrices are limited, recorded levels of BAC (up to 99.6 μg L−1 [15]) and CX (up to 9.57 μg L−1 [14]) in surface water have exceeded those of TCC. Concerns about the potential environmental risks and ecological impact of antimicrobial chemicals in aquatic ecosystems have been raised, as they are frequently detected in surface waters.
Developmental stages are key phases in the life cycles of aquatic organisms, and during their early developmental stages, these organisms are particularly sensitive to environmental contaminants [18]. The impairment of early development may affect the growth and organ development of individual organisms and the health of future generations [18]. Previous studies have found that several antimicrobial chemicals induce developmental toxicity in aquatic organisms at high concentrations. For instance, TCS at 0.5 mg L−1 induces developmental malformation, such as spinal deformities and pericardial edema 96 h post fertilization (hpf) [19]. Furthermore, 225 μg L−1 TCC results in high embryonic mortality, delayed hatching, yolk resorption, and increased malformations [20]. It was also found that 0.05–5 mg L−1 of BAC, BEC, and CX cause zebrafish embryo hatching delay or inhibition and morphological abnormalities and that the acute toxicity of BAC exceeds that of banned TCS and TCC [21]. These studies provide valuable toxicological information on antimicrobial chemicals’ impact on aquatic organism development. The developmental toxicity of antimicrobial chemicals directly reflects their ecological risks and is associated with various biological responses, from genes to tissues [22,23]. However, whether several common antimicrobial chemicals cause developmental toxicity and differences in toxicity to zebrafish embryos at environmentally relevant concentrations remains uncertain.
The mechanisms underlying the developmental toxicity of several of these widely used antimicrobial chemicals to aquatic organisms are currently poorly understood. Prior investigations employing conventional methods that focus on a restricted set of genes and proteins as biological endpoints have failed to fully encompass the comprehensive range of biological responses elicited by the toxicity of antimicrobial chemicals. The interaction between pollutants and endogenous molecules is the cause of toxicity [24]. Pollutants disrupt the normal functions of endogenous signaling molecules, leading to abnormalities in intracellular signaling pathways and thus affecting various physiological processes. The abnormal phenotype of signaling molecules affected by pollutants can be used to evaluate the mechanism of pollutant-induced teratogenicity [25,26]. Nontargeted transcriptome analysis, which comprehensively evaluates gene transcription in cells, is an important emerging tool that can address the above issues. It has been employed to clarify how pollutants adversely affect fish development at the molecular level and determine their modes of action in fish at early life stages [27,28]. Transcriptomic analysis has been used to identify the key genes affected by pollutants and to determine the functional pathways and interaction networks of these genes, thereby revealing the molecular mechanisms of pollutants’ toxic effects [29,30]. The zebrafish (Danio rerio) embryo is a widely used model for studying the developmental toxicity of chemicals because of its sensitivity to toxic substances and the availability of a wide range of developmental parameters that serve as endpoints for assessing embryonic toxicity; examples of these parameters are delayed hatching, heart rate, pericardial edema, yolk sac edema, and morphological abnormalities [24].
In this study, we identified and compared the developmental effects of five widely used antimicrobial chemicals on zebrafish embryos exposed to these chemicals through short-term exposure experiments. Transcriptomic analysis elucidates the underlying molecular mechanisms through which the chemicals adversely affected zebrafish embryo development. This study advances our understanding of the toxicity exerted by antimicrobial chemicals on aquatic organisms and provides novel insights into the underlying mechanisms involved. By investigating developmental toxicity, we contribute to assessing the environmental safety of widely used antimicrobials and their alternatives, ultimately providing a novel perspective on the potential ecological implications of these chemicals.
2. Materials and methods
2.1. Chemicals and reagents
As benzalkonium chloride C12 (BAC-C12; C21H38NCl) is widely used and is the BAC most commonly detected in the environment, it was the target analyte in this study [10,31,32]. This BAC (99 % purity), together with BEC (≥98 % purity) and CX (99 % purity), was purchased from Sigma-Aldrich (St. Louis, MO, USA). Triclosan (99 % purity) and TCC (98 % purity) were purchased from Dr. Ehrenstorfer (Augsburg, Germany). All five antimicrobial chemicals were solubilized in methanol (chromatographic grade) to obtain a master solution placed at 4 °C to ensure antimicrobial solubility and stability, and the resulting solutions were then diluted with dimethyl sulfoxide (DMSO; chromatographic grade) to obtain the desired test concentrations. For more information on the reagents and materials used in this study, refer to the Supporting Information.
2.2. Zebrafish maintenance and embryo collection
Adult wild-type AB zebrafish (approximately six months old) were obtained from the State Key Laboratory of the Chinese Research Academy of Environmental Sciences. The adult zebrafish were maintained in a flow-through system at a water temperature of 27 ± 1 °C and relative humidity of 50 ± 10 % under a photoperiod of 14 h:10 h (light:dark). The dissolved oxygen concentration in water was maintained above 7.72 mg L−1, and the conductivity was 480 μS cm−1. Zebrafish were fed newly hatched brine shrimp (Artemia naupli) twice daily for 14 consecutive days of adaptive feeding. Fertilized eggs were obtained from the natural spawning of adult zebrafish (Danio rerio). The night before the experiment, healthy adult zebrafish were randomly selected and placed into spawning tanks; the bottom of each was equipped with a stainless steel spawning tray covered by a fine net with a mesh size that could be penetrated by eggs. The male and female fish were separated by a partition, and the water temperature was kept at 27 ± 1 °C. The next morning, the clapboard was removed, and the resulting influx of light stimulated spawning. Fertilized eggs were collected within 2 h of spawning in sterile Petri dishes, and healthy embryos with a normally developed blastula were selected under a microscope for exposure experiments [33]. All experiments followed the guidelines for the Care and Use of Laboratory Animals of the Chinese Research Academy of Environmental Sciences.
2.3. Zebrafish embryo exposure design
At 2–4 hpf, 25 blastula-stage embryos per replicate were randomly transferred using clean pipettes to Petri dishes containing 50 mL of an appropriate concentration of an antimicrobial chemical. Three concentrations (0.4, 4, and 40 μg L−1) of each antimicrobial chemical were evaluated to ensure that these amounts in all exposure groups fell within the range of actual environmental water concentrations [10,34]. A solution containing 0.001 % DMSO was used as a blank control, and five replicates of each concentration were used. The embryos were incubated at 28 ± 1 °C, with a 14 h light period followed by a 10 h dark period. The embryos were maintained in a semi-static solution for seven days, with the solution being refreshed every 24 h. The hatchability, mortality, and progression of larvae were assessed at 24 h intervals during the complete exposure period. Daily monitoring included recording the number of deceased embryos and assessing the stage of embryonic development. An embryo without a heartbeat under microscopic observation was considered dead. At 48 hpf, five live embryos from each replicate were selected for heart rate measurement. The number of heartbeats in 20 s was recorded using ZEISS Vert. Axiovert 5 microscope (Jena, Germany). For pigmentation assessment, we followed established protocols in the literature [23,24,35]. We randomly selected five live embryos from each replicate to be visually inspected under a microscope at a consistent angle to assess the degree of pigmentation. Given that zebrafish are three-dimensional, and planar photographs capture only a partial view of the embryo, we based our qualitative scoring on the intensity of pigmentation observed outside the embryo. Five randomly selected live embryos from each replicate were also examined under the same microscope to assess morphological abnormalities, including yolk sac edema, spinal malformations, and pericardial edema, and to calculate the number and incidence of zebrafish presenting with each morphological change. After a seven-day exposure period, the larvae were collected, thoroughly rinsed, and sterilized with phosphate-buffered saline. Next, the larvae were promptly frozen in liquid nitrogen and stored at −80 °C for subsequent experimental analysis.
2.4. RNA isolation, cDNA (Complementary DNA) library construction, and sequencing
The zebrafish larvae were collected and pooled for RNA (Ribonucleic Acid) -seq analysis. Total RNA was extracted from five replicates of each treatment using an RNeasy Mini Kit (Qiagen, Basel, Switzerland). The concentration and purity of the extracted RNA were examined using a Nanodrop 2000 (Thermo Scientific, USA), RNA integrity was assessed by performing agarose gel electrophoresis, and RNA integrity numbers were determined using an Agilent 2100 (Agilent, Santa Clara, CA, USA). A single-cell RNA library was constructed in which the total amount of RNA per cell was at least 1 μg, the concentration of RNA per cell was at least 35 ng μL−1, the optical dispersion at 260 and 280 nm (OD260/280) was at least 1.8, and the OD260/230 was at least 2.0. Sequencing experiments were performed using an Illumina TruSeqM RNA Sample Prep Kit method for library construction, based on an Illumina NovaSeq 6000/HiSeq X Ten (Illumina, USA) sequencing platform. Five replicates per treatment group of all mRNAs transcribed were sequenced, and all mRNAs transcribed were sequenced.
2.5. Transcriptomics sequence (RNA-seq) analysis
First, to ensure the accuracy of subsequent bioinformational analysis, the original sequencing data were filtered using fastp (version 0.19.5) to remove unreliable readings, and the resulting data were subjected to quality assessment to obtain high-quality reads. This quality assessment involved eliminating discarded adapters, sequences with lengths of less than 30 base pairs after mass pruning, ambiguous sequences with an abundance of more than 5 %, and sequences containing over 20 % of low-quality bases (quality value < 10). Table S1 (Supplementary Material) lists the total numbers of raw and clean reads obtained following the quality assessment of each sample. The processed clean reads were sequence aligned with a zebrafish reference genome (version GRCz11; source: http://asia.ensembl.org/Danio_rerio/Info/Index) using HISAT2 (version 2.1.0) to obtain the mapped reads for subsequent transcript assembly and expression calculations. The mapped reads were spliced and assembled using StringTie (version 2.1.2) and then compared with the original genome annotation information for complementation and refinement. RNA-Seq by Expectation Maximization (version 1.3.3) was used to quantitatively analyze the overall expression levels of genes/transcripts. DESeq2 (version 1.24.0) was employed to analyze differentially expressed genes (DEGs) using thresholds of a fold change equal to or greater than 2 and a p-value less than 0.05 [36]. Table S2 (Supplementary Material) lists the total numbers of DEGs (comprising upregulated and downregulated genes) for each treatment. GOATOOLS (version 0.6.5) was used for conducting gene ontology (GO) enrichment analysis on the identified DEGs, while the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology-based Annotation System (version 2.1.1) was employed for performing functional enrichment analysis of KEGG pathways; this was used to distinguish the differentially expressed pathways between the control and treatment groups.
2.6. Real-time quantitative polymerase chain reaction (RT-qPCR) validation
Key DEGs from the enriched pathways were subjected to RT-qPCR to validate the reliability of the transcripts acquired from transcriptomic sequencing. The mRNA primer sequences were obtained from published literature and synthesized by Sangon Biotech (Shanghai, China). Table S3 (Supplementary Material) provides the sequences of the primers. Many studies have demonstrated the stability of β-actin expression in zebrafish by normalizing the expression data of different genes [[37], [38], [39], [40], [41], [42]]; β-actin was thus used as a reference gene [43]. The methods employed to extract and isolate total RNA from each sample were the same as those used in the transcriptomic analysis. Real-time quantitative polymerase chain reaction was carried out in a Fluorescent Quantitative PCR Detection System (LineGene 9600, China) using the SuperReal Premix Plus (SYBR Green) Kit (Tiangen Biotech, Beijing, China). For each gene, qRT-PCR reactions were conducted with three technical replicates for every biological replicate (n = 3). Relative gene expression was calculated using the 2−ΔΔCt method, and the results were compared with transcriptomic sequencing data using linear regression analysis in Origin 2023 (OriginLab, USA) to afford Pearson correlations.
2.7. Data statistics and analysis
Data were graphed and analyzed using GraphPad Prism 9.0 (San Diego, CA, USA) and Origin 2023. Before statistical analysis, the normality of the data was assessed using the Shapiro–Wilk test. The normality test results indicated that the dataset conformed to a normal distribution. Therefore, one-way ANOVA (analysis of variance) was performed to analyze the hatching rate, heart rate, and rates of four typical deformities. Dunnett's test determined the differences between the control and treatment groups. The data on the differences between the various groups are expressed as means ± standard deviations. A p-value less than 0.05 indicated a significant (∗) difference, while a p-value less than 0.01 indicated an extremely significant (∗∗) difference.
3. Results and discussion
3.1. Effects of the five typical antimicrobial chemicals on the hatching rate of zebrafish
Hatching is a key step in the transition from an embryo to a larval fish; therefore, the hatching rate is commonly used to evaluate whether a chemical harms the development and growth of an organism [44]. The hatching rates of the control group at 48, 72, and 96 hpf were 50.0 %, 91.7 %, and 98.3 %, respectively. Compared with the hatching rate of the control group, the hatching rates of the TCS, TCC, and BAC treatment groups were significantly reduced, while the hatching rates of the BEC and CX exposure groups were not significantly reduced (Fig. 1). Triclosan and BAC had the greatest inhibitory effects on hatching. Specifically, there were significantly decreased hatching ratios in all TCS and BAC exposure groups as early as 72 hpf. Among the five tested antimicrobial chemicals, only BAC inhibited hatching at 48 hpf, with an environmentally relevant lowest effective concentration of 0.4 μg L−1. These findings suggest that BAC significantly affects zebrafish embryo developmental processes, particularly hatching. Although the hatching rate does not directly reflect acute toxicity, the results indicate that BAC may be the most potent of the five chemicals regarding developmental toxicity. This is consistent with previous studies reporting significant concentration-dependent hatching inhibition by BAC at higher concentrations (0.1–0.5 mg L−1), in which it also exhibited greater toxicity compared to TCS and TCC [21]. This study is the first to report that short-term exposure to BAC inhibits zebrafish embryo hatching at environmentally relevant concentrations and that BAC exhibits higher toxicity than other widely used disinfectants. These findings highlight the need for more stringent regulation and monitoring of BAC concentrations in aquatic environments to protect organisms. Our results suggest that exposure to certain antimicrobial chemicals may adversely affect zebrafish embryonic development, potentially negatively affecting survival and population size.
Fig. 1.
Chemical structures of triclosan (TCS, a), triclocarban (TCC, b), benzalkonium chloride (BAC, c), benzethonium chloride (BEC, d), and chloroxylenol (CX, e), along with their effects on the hatching rates of zebrafish embryos at 48, 72, and 96 h post-fertilization (hpf). Data shown as mean ± standard deviation. Asterisks indicate significant differences from control (∗p < 0.05; ∗∗p < 0.01; Dunnett's test).
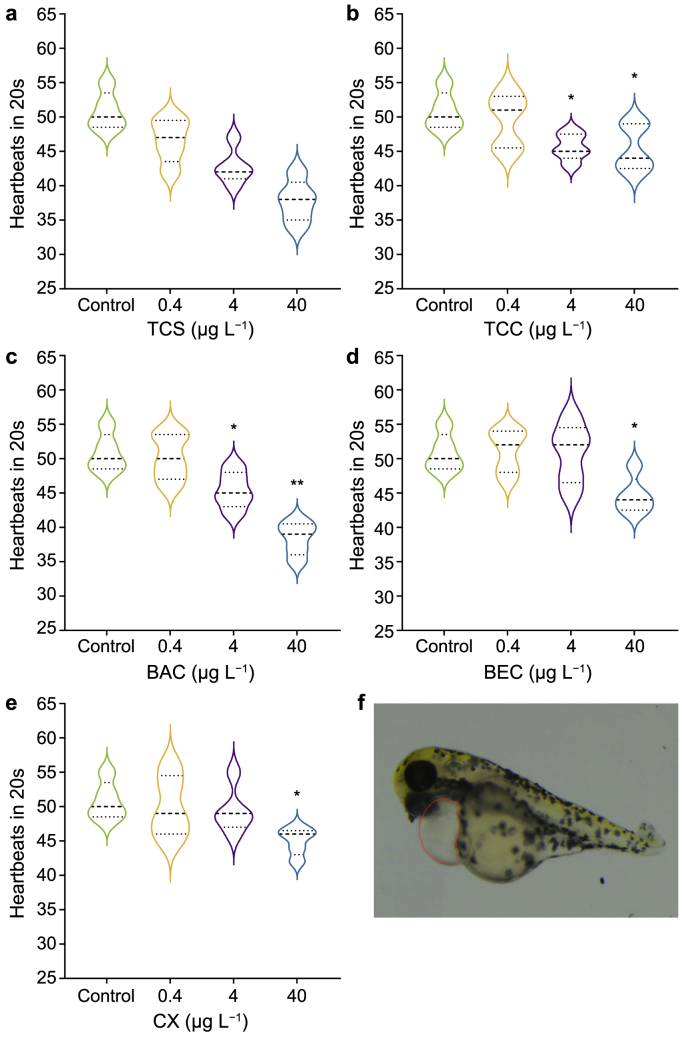
3.2. Effect of typical antimicrobial chemicals on the heart rate of zebrafish
Alterations in zebrafish embryo heart rates during hatching were further evaluated. Significant dose–response decreases in heart rate were observed at 48 hpf in embryos exposed to the chemicals, with the lowest effective concentrations being 4 μg L−1 for TCS, TCC, and BAC and 40 μg L−1 for BEC and CX (Fig. 2). Compared with the two other replaced chemicals, namely, CX and BEC, BAC had a greater effect on the heart rate of the embryos. At an exposure concentration of 4 μg L−1, the BAC-treated group showed an 89.4 % reduction in heart rate compared with the control group. This reduction was similar to those observed in the groups treated with the banned chemicals TCS and TCC, which were 84.3 % and 89.8 %, respectively. This result indicates that BAC, TCS, and TCC are similarly toxic to embryos regarding heart rate. Consistent with the hatching rate results, BEC and CX had a lower heart rate impact than TCS, TCC, and BAC. At a concentration of 40 μg L−1, BEC and CX reduced heart rates by 87.8 % and 88.6 %, respectively. By contrast, TCS, TCC, and BAC induced similar adverse effects at a much lower concentration of 4 μg L−1. The heart is the first organ to form and function during zebrafish development, and the heart rate is an important indicator of heart function [45]. Thus, the significant decreases in heart rate caused by exposure to these chemicals indicate that they can negatively affect the development and function of the heart in the early developmental stages of zebrafish, leading to developmental defects and phenotypic abnormalities in zebrafish embryos [46].
Fig. 2.
a–e, Heart rate (number of heartbeats in 20s) of zebrafish embryos at 48 h post-fertilization after exposure to triclosan (TCS, a), triclocarban (TCC, b), benzalkonium chloride (BAC, c), benzethonium chloride (BEC, d), and chloroxylenol (CX, e). Box plots show median (center line) and interquartile range (25th-75th percentiles). Asterisks denote significant differences between treatments and the control (determined by a Dunnett post hoc comparison; results are shown as means ± standard deviations (∗p < 0.05, ∗∗p < 0.01). f, Location of zebrafish heart (red circle).
3.3. Toxic effects of antimicrobial chemicals on the early development of zebrafish
Damage or disruption to the heart may affect the overall health and development of zebrafish [46]. Since embryo heart rates decreased at 48 hpf because of antimicrobial chemical exposure, further evaluations were conducted to assess whether there were any developmental or other physiological defects in the treatment groups compared with the control group. Developmental toxicity tests showed that treatment with TCS, TCC, BAC, or BEC, but not CX, had significant teratogenic effects in zebrafish. For example, spinal deformation was observed in the groups treated with TCS, TCC, BAC, and BEC (Fig. 3), with the lowest effective concentrations being 4 μg L−1 for TCC and 40 μg L−1 for TCS, BAC, and BEC. These results indicate that such chemicals may perturb the early skeletal development stages of zebrafish [23], leading to the development of spinal deformities. Although BAC and BEC caused spinal deformities at an effective concentration of 40 μg L−1, the deformity rate for the BAC and BEC treatments was much lower than that for the TCS and TCC treatments. Moreover, unlike BAC, BEC, and CX, TCS and TCC induced yolk sac edema in zebrafish. The effective concentration of TCS was 4 μg L−1, significantly lower than that of TCC (i.e., 40 μg L−1). This result indicates that TCS was more toxic to zebrafish yolk sacs than TCC was. As the yolk sac serves as the nutrient reserve organ for the zebrafish embryo [35], yolk sac abnormities induced by TCS and TCC may affect the nutrient supply during zebrafish development.
Fig. 3.
a–e, Developmental abnormalities in zebrafish embryos exposed to antimicrobial compounds. Incidence rates of four typical deformities (yolk sac edema, spine deformation, and pericardial edema at 72 h post-fertilization (hpf) and pigmentation inhibition at 48 hpf) following exposure to triclosan (TCS, a), triclocarban (TCC, b), benzalkonium chloride (BAC, c), benzethonium chloride (BEC, d), and chloroxylenol (CX, e). Results are presented as means ± standard deviations. Significant differences versus the control are indicated as ∗p < 0.05, ∗∗p < 0.01 (ANOVA, Dunnett's test). f, Representative brightfield images showing the teratogenic effects of antimicrobial chemicals at 72 hpf. yse: yolk sac edema; sd: spine deformation; pe: pericardial edema.
Furthermore, TCS and TCC had different teratogenic effects on zebrafish. Specifically, treatment with 40 μg L−1 TCS inhibited pigment deposition in zebrafish, while treatment with the same concentration of TCC caused pericardial edema (Fig. 3a and b). This result suggests that these two chemicals may have different negative impacts on the health and survival of zebrafish during development. By contrast, treatment with CX at various concentrations had no teratogenic effects, which may be attributable to the structure of CX or the treatment concentrations. Additionally, the occurrence of multiple teratogenic changes in single fish suggests that some of these antimicrobial chemicals may exhibit broad-spectrum toxicity. That is, they adversely affect the normal development of zebrafish embryos in multiple ways. The results of the hatching rate and teratogenic tests indicate that exposure to antimicrobial chemicals adversely affects the early growth and development of zebrafish. However, the potential molecular mechanisms driving these downstream effects remain poorly understood. We employed transcriptomic analysis to clarify the molecular mechanisms of these chemicals’ toxic effects during zebrafish development.
3.4. Transcriptomic analysis of zebrafish larvae
We performed RNA sequencing-based transcriptomic analysis of larvae that survived exposure to the antimicrobial chemicals to explore the potential molecular mechanisms underlying the observed developmental and physiological defects caused by exposure. After adaptor trimming and quality control, 520.62 Gb of clean reads were obtained. The clean reads of all samples totaled more than 6.01 Gb, with a Q20 percentage of over 96.46 % and a Q30 percentage of over 90.67 %. Sequence comparisons of the clean reads of each sample with the genome of zebrafish (Danio rerio) were conducted, and the matching rate ranged from 88.73 % to 93.98 %. This analysis detected 34,890 genes; 28,681 of these genes were known, and 6209 had not been previously characterized.
Volcano plots were drawn based on the log2-fold changes in transcript expression in larvae and their corresponding −log 10 significance values. These plots show the overall scattering of various transcripts and the upregulated and downregulated genes in each treatment group (Supplementary Material Fig. S1). There were dose-dependent increases in the numbers of DEGs in the groups treated with the five antimicrobial chemicals. The group treated with 40 μg L−1 of TCC had the most DEGs, with 3726 being upregulated and 399 being downregulated. The groups treated with 0.4, 4, and 40 μg L−1 of BEC had relatively high numbers of DEGs, 1,823, 1,988, and 1,932, respectively (Supplementary Material Table S2). A Venn diagram was drawn to illustrate the relationships between DEGs and the overlapping of DEGs in the groups treated with the five antimicrobial chemicals, revealing that 13,642 genes were differentially expressed in all groups (Supplementary Material Fig. S2a). Moreover, principal component analysis (Supplementary Material Fig. S2b) showed that the five antimicrobial chemicals had similar effects at the transcriptional level, indicating that they may have similar modes of action. Pathway annotation analysis of these common DEGs using KEGG revealed that they are typically involved in metabolic pathways associated with five categories: metabolism, environmental information processing, cellular processes, organismal systems, and human diseases. The KEGG annotations of these DEGs indicate that the five antimicrobial chemicals primarily affect the immune system and signal transduction functions in the zebrafish brain (Supplementary Material Fig. S3). The DEGs for each processing group were also functionally annotated using the GO classification standard. The GO enrichment analysis (Supplementary Material Fig. S4) revealed that the DEGs across all treatment groups were predominantly associated with cellular processes, biological regulation, and metabolism. These categories encompass extracellular structure organization, extracellular matrix organization, negative regulation of hydrolase activity, and regulation of endopeptidase activity.
We performed a systematic KEGG functional enrichment analysis to clarify the underlying mechanisms. The top 20 enriched KEGG pathways (with the most DEGs or minimum p-adjust values) were obtained (Fig. 4), three associated with the digestive system, three with xenobiotic biodegradation and metabolism, and three with lipid metabolism. Detailed information on the signaling pathways, categories, related genes, and numbers of DEGs can be found in Table S4 (Supplementary Material). Fig. 4a shows that the signaling pathways highly enriched in zebrafish treated with the antimicrobial chemicals were the complement and coagulation cascade pathway and the mitogen-activated protein kinase (MAPK) signaling pathway. The MAPK signaling pathway is central to the embryonic development of vertebrates, as it aids in the regulation of hormones and in immune response and contributes to overall growth [47,48]. Similarly, the complement and coagulation cascade pathway is involved in immune response. It plays an important role in antimicrobial defense reactions, immune regulation, and mediation of the damaging reactions of immune pathologies [49]. The observed developmental abnormalities may be linked to disruptions in these significantly enriched pathways in the treated zebrafish. However, further investigation is required to establish a more direct relationship between pathway disruptions and the toxic phenotypes, such as malformations, observed in this study.
Fig. 4.
a, A systematic Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis showing top 20 enriched pathways based on differentially expressed genes (DEGs) or the lowest p-adjust values. ECM: extracellular matrix; PPAR: peroxisome proliferator-activated receptor. b, Protein-protein interaction network of hub genes and their interacting genes in the complement and coagulation cascades pathway and mitogen-activated protein kinase (MAPK) signaling pathway. Node size indicates connectivity degree. Circular nodes represent transcription factors; triangular nodes represent non-transcription factor proteins.
To explore the interactions between DEGs, we used the search tool of the Retrieval of Interacting Genes/Proteins Database and Cytoscape to conduct a protein–protein interaction (PPI) network analysis of genes encoding proteins in the MAPK signaling pathway and the complement and coagulation cascade pathway, respectively. We then generated a gene expression network. Fig. 4b shows that the complement and coagulation cascade pathway network comprises 55 nodes connected by 100 edges. Five hub genes were identified, namely f2, cfb, c5, c9, and plg, and these may modulate the complement and coagulation cascade pathways. The MAPK signaling pathway network consists of 21 nodes connected by 61 edges. Four hub genes were identified, namely, mapk14a, mapk14b, frk, and jun, which are the core nodes of the MAPK signaling pathway. The network analysis results show that mapk14a and frk share a large number of co-regulatory genes with mapk14b. These hub genes play crucial roles in connecting and regulating the complex signaling pathway network, thereby affecting the network's activity and stability.
3.5. RT-qPCR validation of hub genes for the selected key pathways
The biological pathways and interaction networks associated with hub genes are important components of toxic processes. Thus, the expression levels of hub genes in the complement coagulation cascade pathway and the MAPK pathway were examined. The aim was to verify the interactions and functions of these genes in biological processes and thereby reveal the molecular mechanisms underpinning the developmental toxicity of antimicrobial chemicals in zebrafish.
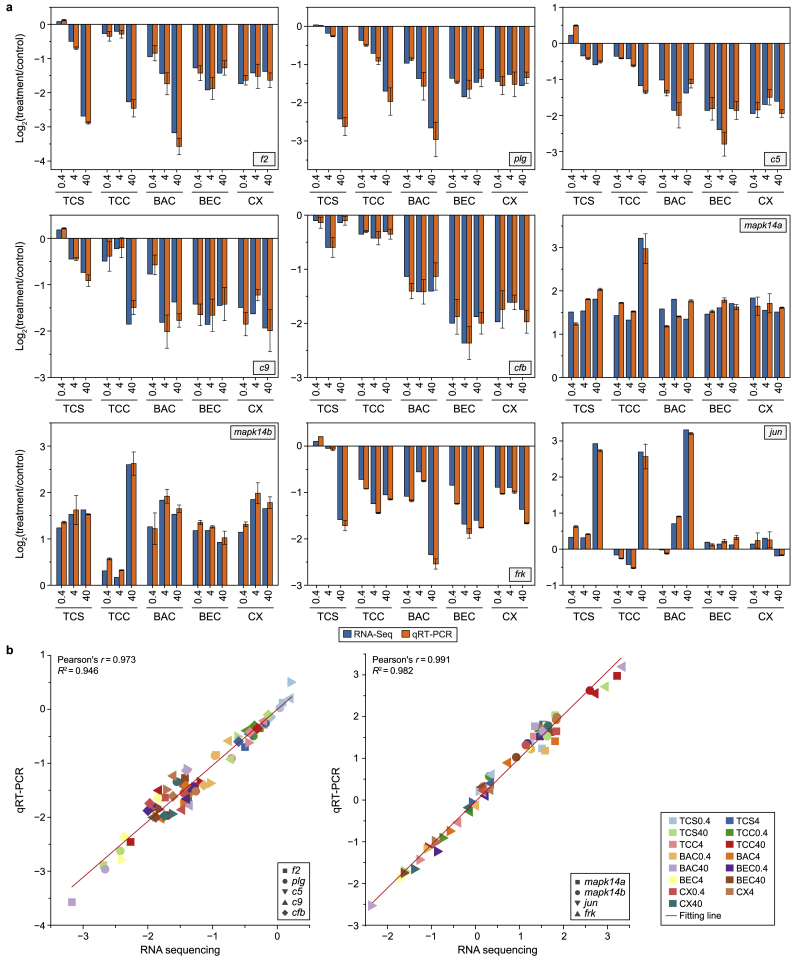
First, the five hub genes in the PPI network of the complement and coagulation cascade pathways were subjected to RT-qPCR validation. In general, the expression levels and regulatory trends of the genes detected using RNA-seq were consistent with the results obtained from qualification by RT-qPCR in all five chemical exposure groups (Fig. 5a). Relative to the control, the transcripts of f2 (1.17- and 3.17-fold) and plg (1.42- and 2.66-fold) were significantly downregulated (p < 0.01) in the high-concentration treatment groups (40 μg L−1). f2 and plg encode two important proteins of the coagulation cascade pathway. f2 encodes coagulation factor II, which is activated to convert prothrombin into thrombin, thereby promoting coagulation [50]. Thrombin is an essential link between inflammation and hemostasis, key processes in the body's defense against infection [51]. plg encodes plasminogen, a precursor protein involved in the fibrinolytic system. Plasminogen is converted into plasmin, an enzyme that plays an important role in the breakdown of blood clots [[52], [53], [54]]. A reduction in the expression of f2 and plg induced by antimicrobial exposure may lead to a decrease in thrombin generation and impaired plasminogen function in the early development of zebrafish. This could contribute to developing inflammatory diseases and potentially lead to late-onset mortality in zebrafish [55]. The expression of the classical complement component-related genes c5, c9, and cfb was significantly downregulated (p < 0.01) in the groups treated with high concentrations (40 μg L−1) of the five antimicrobial chemicals (Fig. 5a). Relative to the control, the expression of c5 was significantly downregulated (p < 0.05) in the groups treated with 40 μg L−1 of TCS (−0.52-fold), TCC (−1.36-fold), BAC (−1.12-fold), BEC (−1.86-fold), and CX (−1.94-fold). Similarly, relative to the control, the expression of c9 was significantly downregulated (p < 0.01) in the groups treated with 40 μg L−1 of TCS (−0.91-fold), TCC (−1.49-fold), BAC (−1.77-fold), BEC (−1.42-fold), and CX (−1.99-fold). The complement system is an indispensable part of the immune response, as it plays an important role in supporting resistance to infection and regulating immune balance [56]. Animals lacking various complement components exhibit a range of phenotypes associated with suboptimal host defense, including reduced phagocytic activity, increased susceptibility to infection, impaired T and B cell responses, and other compromised immune capabilities [49]. The results of this study show that exposure to antimicrobial chemicals significantly reduces c5 and c9 expression, suggesting that these chemicals may impair immune function in fish. cfb encodes complement factor B, a crucial protein in the complement system that plays a key role in immune response by regulating inflammatory and immunomodulatory processes [57,58]. A reduced expression of cfb may result in the inadequate synthesis of complement factor B, thereby affecting immune regulation.
Fig. 5.
a, Validation of RNA sequencing data by quantitative real-time polymerase chain reaction (qRT-PCR). The expression of selected target genes, shown as log2-fold changes, was consistent with RNA sequencing results across exposure treatments compared to the control. TCS: triclosan; TCC: triclocarban; BAC: benzalkonium chloride; BEC: benzethonium chloride; CX: chloroxylenol. b, Correlation analysis between RNA sequencing (x-axis) and qRT-PCR data (y-axis) for genes in the complement and coagulation cascades and mitogen-activated protein kinase (MAPK) signaling pathways. Pearson correlation analysis revealed significant positive correlations (p < 0.01) between the two datasets.
Second, the four hub genes in the PPI network of the MAPK signaling pathway were subjected to RT-qPCR analysis (Fig. 5). It was found that exposure to the five antimicrobial chemicals significantly upregulated mapk14a and mapk14b expression compared with the control group. mapk14a (full name: mitogen-activated protein kinase 14a; also known as p38α) and mapk14b (full name: mitogen-activated protein kinase 14b; also known as p38β) encodes p38, which is a member of the MAPK family. The activation of mapk14a and mapk14b is usually associated with apoptosis, and their corresponding proteins regulate the function of downstream target proteins through phosphorylation [[59], [60], [61]], thereby affecting embryonic development [62]. Thus, the increased expression levels of mapk14a and mapk14b in zebrafish treated with the five antimicrobials may be associated with the observed malformation of zebrafish larvae. Mapk14a regulates heart formation, differentiation, and maturation during zebrafish development, playing a crucial signaling role in cardiac development. Changes in its expression can lead to heart defects, including structural abnormalities and issues with cardiomyocyte proliferation and differentiation. Additionally, mapk14a is involved in the heart's adaptive response to environmental stressors, such as chemical pollutants or drug exposure [62]. Dysregulation of Mapk14a may be associated with heart developmental defects in zebrafish [60]. Activated mapk14a was previously shown to block the proliferation of adult zebrafish cardiomyocyte proliferation [63]. Therefore, the significant decrease in the heart rates of zebrafish larvae following treatment with the five antimicrobial chemicals may be attributable to the substantial upregulation of mapk14a experssion. By contrast, compared to the control, frk expression was significantly downregulated (p < 0.01) upon exposure to 40 μg L−1 of TCS (−1.59-fold), TCC (−1.05-fold), BAC (−2.34-fold), BEC (−1.60-fold), and CX (−1.36-fold) (Fig. 5). The fyn-related kinase (frk) protein encoded by frk regulates cell growth, proliferation, and survival. It is involved in MAPK signaling pathway activation [64]. Frk gene expression regulates osteoblast differentiation and bone growth during zebrafish skeletal development, playing a crucial role in bone morphogenesis and development [65]. This protein also significantly impacts the embryonic and craniofacial development of zebrafish [65]. Therefore, the downregulation of frk expression by exposure to antimicrobial chemicals may lead to developmental deficits and malformation in zebrafish larvae, affecting their overall health and development. Compared with jun expression in the groups treated with 40 μg L−1 of BEC and CX, respectively, jun expression was significantly increased in the groups treated with 40 μg L−1 of BAC, TCS, and TCC. Jun (also commonly referred to as c-Jun) is a key component of the MAPK pathway that is activated by phosphorylation and mediates apoptosis [66,67]. Therefore, the significantly increased expression of jun in larvae induced by BAC, TCS, or TCC exposure might have led to an increase in cell apoptosis. However, this finding needs further experimental verification. In line with the above-described results, bisphenol A was previously shown to induce apoptosis in zebrafish by affecting the expression of jun [68]. Overall, our transcriptomic sequencing findings established the relationship between the developmental toxicity of zebrafish larvae treated with the five antimicrobial chemicals and the coagulation cascade and MAPK signaling pathways, with these relationships reflected in the morphological results.
Furthermore, the correlations between the sequencing results of the hub genes in the complement and coagulation cascade pathway and the MAPK pathway and the results of RT-qPCR were analyzed, and their statistical significance was determined by performing Pearson correlation tests. Fig. 5b shows the correlation data of the hub genes detected by RNA sequencing and RT-qPCR. The results show that the qualitative consistencies between the upregulated and downregulated genes in the complement and coagulation cascade pathway (R2 = 0.946) and between those in the MAPK pathway (R2 = 0.982) were strong, indicating that the correlations between RNA sequencing data and RT-qPCR data were highly statistically significant.
4. Conclusion
In this study, we compared the development toxicological effects of five common antimicrobial chemicals on zebrafish during their early life stages and explored the potential mechanisms underlying these effects using transcriptomic analysis. The zebrafish embryos were more adversely affected by the replacement antimicrobial chemical BAC than by the banned chemicals TCS and TCC, exhibiting delayed or inhibited hatching, embryo death, reduced heart rate, and developmental morphology deformities.
By contrast, zebrafish were less affected by the BEC and CX replacement antimicrobial chemicals than BAC, TCS, and TCC. Transcriptomic analyses revealed that the interactions between hub genes in the MAPK signaling and coagulation cascade pathways may be crucial in influencing zebrafish development. The downregulation of hub genes in the coagulation cascade pathway weakens immune function, while the upregulation of hub genes in the MAPK signaling pathway may trigger cell apoptosis, potentially contributing to increased mortality. These molecular alterations suggest a mechanistic link to developmental toxicity.
Our findings comprehensively compare the adverse effects of five commonly used antimicrobial chemicals on zebrafish development, offering a possible mechanistic explanation for their developmental toxicities. These results highlight the potential environmental risks these chemicals pose to aquatic ecosystems.
CRediT authorship contribution statement
Yueyue Liu: Writing – review & editing, Writing – original draft, Software, Methodology, Formal analysis, Data curation. Chen Wang: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Zhiyou Fu: Methodology, Formal analysis. Yingchen Bai: Formal analysis, Data curation. Guomao Zheng: Methodology, Data curation. Fengchang Wu: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful to the National Natural Science Foundation of China (42394154, 42394152, 42377274) for providing financial support for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2025.100543.
Contributor Information
Chen Wang, Email: wangchen@craes.org.cn.
Fengchang Wu, Email: wufengchang@vip.skleg.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Hora P.I., Pati S.G., McNamara P.J., Arnold W.A. Increased use of quaternary ammonium compounds during the SARS-CoV-2 pandemic and beyond: consideration of environmental implications. Environ. Sci. Technol. Lett. 2020;7(9):622–631. doi: 10.1021/acs.estlett.0c00437. [DOI] [PubMed] [Google Scholar]
- 2.Zhang C., Cui F., Zeng G.M., Jiang M., Yang Z.Z., Yu Z.G., Zhu M.Y., Shen L.Q. Quaternary ammonium compounds (QACs): a review on occurrence, fate and toxicity in the environment. Sci. Total Environ. 2015;518–519:352–362. doi: 10.1016/j.scitotenv.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Rundle C.W., Hu S., Presley C.L., Dunnick C.A. Triclosen and its alternatives in antibacterial soaps. Dermatitis. 2019;30(6):352–357. doi: 10.1097/DER.0000000000000519. [DOI] [PubMed] [Google Scholar]
- 4.Safety and effectiveness of consumer antiseptics; topical antimicrobial drug products for over-the-counter human use. Final rule. Fed. Regist. 2016;81(172):61106–61130. [PubMed] [Google Scholar]
- 5.United States Environmental Protection Agency, About List N: Disinfectants for Coronavirus (COVID-19). In.
- 6.Adhikari S., Kumar R., Driver E.M., Perleberg T.D., Yanez A., Johnston B., Halden R.U. Mass trends of parabens, triclocarban and triclosan in Arizona wastewater collected after the 2017 FDA ban on antimicrobials and during the COVID-19 pandemic. Water Res. 2022;222 doi: 10.1016/j.watres.2022.118894. [DOI] [PubMed] [Google Scholar]
- 7.Belova L., Poma G., Roggeman M., Jeong Y., Kim D.-H., Berghmans P., Peters J., Salamova A., van Nuijs A.L.N., Covaci A. Identification and characterization of quaternary ammonium compounds in Flemish indoor dust by ion-mobility high-resolution mass spectrometry. Environ. Int. 2023;177 doi: 10.1016/j.envint.2023.108021. [DOI] [PubMed] [Google Scholar]
- 8.Zheng G., Filippelli G.M., Salamova A. Increased indoor exposure to commonly used disinfectants during the COVID-19 pandemic. Environ. Sci. Technol. Lett. 2020;7(10):760–765. doi: 10.1021/acs.estlett.0c00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng G., Webster T.F., Salamova A. Quaternary ammonium compounds: bioaccumulation potentials in humans and levels in blood before and during the covid-19 pandemic. Environ. Sci. Technol. 2021;55(21):14689–14698. doi: 10.1021/acs.est.1c01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S., Ji K., Shin H., Park S., Kho Y., Park K., Kim K., Choi K. Occurrences of benzalkonium chloride in streams near a pharmaceutical manufacturing complex in Korea and associated ecological risk. Chemosphere. 2020;256 doi: 10.1016/j.chemosphere.2020.127084. [DOI] [PubMed] [Google Scholar]
- 11.Olkowska E., Polkowska Ż., Namieśnik J. A solid phase extraction-ion chromatography with conductivity detection procedure for determining cationic surfactants in surface water samples. Talanta. 2013;116:210–216. doi: 10.1016/j.talanta.2013.04.083. [DOI] [PubMed] [Google Scholar]
- 12.Li X., Brownawell B.J. Quaternary ammonium compounds in urban estuarine sediment environments - a class of contaminants in need of increased attention? Environ. Sci. Technol. 2010;44(19):7561–7568. doi: 10.1021/es1011669. [DOI] [PubMed] [Google Scholar]
- 13.Mulder I., Siemens J., Sentek V., Amelung W., Smalla K., Jechalke S. Quaternary ammonium compounds in soil: implications for antibiotic resistance development. Rev. Environ. Sci. Biotechnol. 2018;17(1):159–185. [Google Scholar]
- 14.Tan J., Kuang H., Wang C., Liu J., Pang Q., Xie Q., Fan R. Human exposure and health risk assessment of an increasingly used antibacterial alternative in personal care products: chloroxylenol. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147524. [DOI] [PubMed] [Google Scholar]
- 15.Liao M., Wei S., Zhao J., Wang J., Fan G. Risks of benzalkonium chlorides as emerging contaminants in the environment and possible control strategies from the perspective of ecopharmacovigilance. Ecotoxicol. Environ. Saf. 2023;266 doi: 10.1016/j.ecoenv.2023.115613. [DOI] [PubMed] [Google Scholar]
- 16.Halden R.U., Paull D.H. Co-occurrence of triclocarban and triclosan in U.S. Water resources. Environ. Sci. Technol. 2005;39(6):1420–1426. doi: 10.1021/es049071e. [DOI] [PubMed] [Google Scholar]
- 17.Dhillon G.S., Kaur S., Pulicharla R., Brar S.K., Cledón M., Verma M., Triclosan R. Y. Surampalli. Current status, occurrence, environmental risks and bioaccumulation potential. Int. J. Environ. Res. Publ. Health. 2015;12(5):5657–5684. doi: 10.3390/ijerph120505657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barros S., Alves N., Pinheiro M., Ribeiro M., Morais H., Montes R., Rodil R., Quintana J.B., Coimbra A.M., Santos M.M., Neuparth T. Are fish populations at risk? Metformin disrupts zebrafish development and reproductive processes at chronic environmentally relevant concentrations. Environ. Sci. Technol. 2023;57(2):1049–1059. doi: 10.1021/acs.est.2c05719. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira R., Domingues I., Koppe Grisolia C., Soares A.M. Effects of triclosan on zebrafish early-life stages and adults. Environ. Sci. Pollut. Res. Int. 2009;16(6):679–688. doi: 10.1007/s11356-009-0119-3. [DOI] [PubMed] [Google Scholar]
- 20.Xu J., Qian Q., Xia M., Wang X., Wang H. Trichlorocarban induces developmental and immune toxicity to zebrafish (Danio rerio) by targeting TLR4/MyD88/NF-κB signaling pathway. Environ. Pollut. 2021;273 doi: 10.1016/j.envpol.2021.116479. [DOI] [PubMed] [Google Scholar]
- 21.Sreevidya V.S., Lenz K.A., Svoboda K.R., Ma H. Benzalkonium chloride, benzethonium chloride, and chloroxylenol - three replacement antimicrobials are more toxic than triclosan and triclocarban in two model organisms. Environ. Pollut. 2018;235:814–824. doi: 10.1016/j.envpol.2017.12.108. [DOI] [PubMed] [Google Scholar]
- 22.Robinson J.F., Pennings J.L.A., Piersma A.H. In: Developmental Toxicology: Methods and Protocols. Harris C., Hansen J.M., editors. Humana Press; Totowa, NJ: 2012. A review of toxicogenomic approaches in developmental toxicology; pp. 347–371. [DOI] [PubMed] [Google Scholar]
- 23.He J.H., Gao J.M., Huang C.J., Li C.Q. Zebrafish models for assessing developmental and reproductive toxicity. Neurotoxicol. Teratol. 2014;42:35–42. doi: 10.1016/j.ntt.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Chen J.B., Gao H.W., Zhang Y.L., Zhang Y., Zhou X.F., Li C.Q., Gao H.P. Developmental toxicity of diclofenac and elucidation of gene regulation in zebrafish (Danio rerio) Sci. Rep. 2014;4:4841. doi: 10.1038/srep04841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sipes N.S., Padilla S., Knudsen T.B. Zebrafish: as an integrative model for twenty-first century toxicity testing. Birth defects research. Part C, Embryo. Today Off. : reviews. 2011;93(3):256–267. doi: 10.1002/bdrc.20214. [DOI] [PubMed] [Google Scholar]
- 26.McCollum C.W., Ducharme N.A., Bondesson M., Gustafsson J.A. Developmental toxicity screening in zebrafish. Birth defects research. Part C, Embryo today : reviews. 2011;93(2):67–114. doi: 10.1002/bdrc.20210. [DOI] [PubMed] [Google Scholar]
- 27.Tom M., Auslander M. Transcript and protein environmental biomarkers in fish--a review. Chemosphere. 2005;59(2):155–162. doi: 10.1016/j.chemosphere.2004.10.063. [DOI] [PubMed] [Google Scholar]
- 28.Rogers E.D., Henry T.B., Twiner M.J., Gouffon J.S., McPherson J.T., Boyer G.L., Sayler G.S., Wilhelm S.W. Global gene expression profiling in larval zebrafish exposed to microcystin-LR and microcystis reveals endocrine disrupting effects of Cyanobacteria. Environ. Sci. Technol. 2011;45(5):1962–1969. doi: 10.1021/es103538b. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Zhou Q., Zou W., Hu X. Molecular mechanisms of developmental toxicity induced by graphene oxide at predicted environmental concentrations. Environ. Sci. Technol. 2017;51(14):7861–7871. doi: 10.1021/acs.est.7b01922. [DOI] [PubMed] [Google Scholar]
- 30.Peng Y., Fang W., Yan L., Wang Z., Wang P., Yu J., Zhang X. Early life stage bioactivity assessment of short-chain chlorinated paraffins at environmentally relevant concentrations by concentration-dependent transcriptomic analysis of zebrafish embryos. Environ. Sci. Technol. 2020;54(2):996–1004. doi: 10.1021/acs.est.9b04879. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Carballo E., Sitka A., González-Barreiro C., Kreuzinger N., Fürhacker M., Scharf S., Gans O. Determination of selected quaternary ammonium compounds by liquid chromatography with mass spectrometry. Part I. Application to surface, waste and indirect discharge water samples in Austria. Environ. Pollut. 2007;145(2):489–496. doi: 10.1016/j.envpol.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Butkovskyi A., Leal L.H., Zeeman G., Rijnaarts H.H.M. Micropollutants in source separated wastewater streams and recovered resources of source separated sanitation. Environ. Res. 2017;156:434–442. doi: 10.1016/j.envres.2017.03.044. [DOI] [PubMed] [Google Scholar]
- 33.Babić S., Barišić J., Stipaničev D., Repec S., Lovrić M., Malev O., Martinović-Weigelt D., Čož-Rakovac R., Klobučar G. Assessment of river sediment toxicity: combining empirical zebrafish embryotoxicity testing with in silico toxicity characterization. Sci. Total Environ. 2018;643:435–450. doi: 10.1016/j.scitotenv.2018.06.124. [DOI] [PubMed] [Google Scholar]
- 34.Ramaswamy B.R., Shanmugam G., Velu G., Rengarajan B., Larsson D.G. GC-MS analysis and ecotoxicological risk assessment of triclosan, carbamazepine and parabens in Indian rivers. J. Hazard Mater. 2011;186(2–3):1586–1593. doi: 10.1016/j.jhazmat.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 35.Mu X., Huang Y., Li X., Lei Y., Teng M., Li X., Wang C., Li Y. Developmental effects and estrogenicity of bisphenol A alternatives in a zebrafish embryo model. Environ. Sci. Technol. 2018;52(5):3222–3231. doi: 10.1021/acs.est.7b06255. [DOI] [PubMed] [Google Scholar]
- 36.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 37.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y., Xie S., Yao J. Identification of novel reference genes suitable for qRT-PCR normalization with respect to the zebrafish developmental stage. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filby A.L., Tyler C.R. Appropriate 'housekeeping' genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol. Biol. 2007;8:10. doi: 10.1186/1471-2199-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCurley A.T., Callard G.V. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang R., Dodd A., Lai D., McNabb W.C., Love D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. 2007;39(5):384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadei R., Pelleri M.C., Vitale L., Facchin F., Lenzi L., Canaider S., Strippoli P., Frabetti F. Identification of housekeeping genes suitable for gene expression analysis in the zebrafish. Gene Expr. Patterns : GEP. 2011;11(3–4):271–276. doi: 10.1016/j.gep.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Chen H., Qiu W., Yang X., Chen F., Chen J., Tang L., Zhong H., Magnuson J.T., Zheng C., Xu E.G. Perfluorooctane sulfonamide (PFOSA) induces cardiotoxicity via aryl hydrocarbon receptor activation in zebrafish. Environ. Sci. Technol. 2022;56(12):8438–8448. doi: 10.1021/acs.est.1c08875. [DOI] [PubMed] [Google Scholar]
- 44.Ren X., Zhang H., Geng N., Xing L., Zhao Y., Wang F., Chen J. Developmental and metabolic responses of zebrafish (Danio rerio) embryos and larvae to short-chain chlorinated paraffins (SCCPs) exposure. Sci. Total Environ. 2018;622–623:214–221. doi: 10.1016/j.scitotenv.2017.11.304. [DOI] [PubMed] [Google Scholar]
- 45.Krzykwa J.C., Olivas A., Sellin Jeffries M.K. Development of cardiovascular and neurodevelopmental metrics as sublethal endpoints for the Fish embryo toxicity test. Environ. Toxicol. Chem. 2018;37(10):2530–2541. doi: 10.1002/etc.4212. [DOI] [PubMed] [Google Scholar]
- 46.Xia S., Zhu X., Yan Y., Zhang T., Chen G., Lei D., Wang G. Developmental neurotoxicity of antimony (Sb) in the early life stages of zebrafish. Ecotoxicol. Environ. Saf. 2021;218 doi: 10.1016/j.ecoenv.2021.112308. [DOI] [PubMed] [Google Scholar]
- 47.Yan L., Carr J., Ashby P.R., Murry-Tait V., Thompson C., Arthur J.S. Knockout of ERK5 causes multiple defects in placental and embryonic development. BMC Dev. Biol. 2003;3:11. doi: 10.1186/1471-213X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park J., An G., Park H., Hong T., Lim W., Song G. Developmental defects induced by thiabendazole are mediated via apoptosis, oxidative stress and alteration in PI3K/Akt and MAPK pathways in zebrafish. Environ. Int. 2023;176 doi: 10.1016/j.envint.2023.107973. [DOI] [PubMed] [Google Scholar]
- 49.Satyam A., Graef E.R., Lapchak P.H., Tsokos M.G., Dalle Lucca J.J., Tsokos G.C. Complement and coagulation cascades in trauma. Acute Medicine & Surgery. 2019;6(4):329–335. doi: 10.1002/ams2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Demirci F.Y., Dressen A.S., Kammerer C.M., Barmada M.M., Kao A.H., Ramsey-Goldman R., Manzi S., Kamboh M.I. Functional polymorphisms of the coagulation factor II gene (F2) and susceptibility to systemic lupus erythematosus. J. Rheumatol. 2011;38(4):652–657. doi: 10.3899/jrheum.100728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cicala C., Cirino G. Linkage between inflammation and coagulation: an update on the molecular basis of the crosstalk. Life Sci. 1998;62(20):1817–1824. doi: 10.1016/s0024-3205(97)01167-3. [DOI] [PubMed] [Google Scholar]
- 52.Silva L.M., Doyle A.D., Greenwell-Wild T., Dutzan N., Tran C.L., Abusleme L., Juang L.J., Leung J., Chun E.M., Lum A.G., Agler C.S., Zuazo C.E., Sibree M., Jani P., Kram V., Martin D., Moss K., Lionakis M.S., Castellino F.J., Kastrup C.J., Flick M.J., Divaris K., Bugge T.H., Moutsopoulos N.M. Fibrin is a critical regulator of neutrophil effector function at the oral mucosal barrier. Science (New York, N.Y.) 2021;374(6575) doi: 10.1126/science.abl5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bugge T.H., Flick M.J., Daugherty C.C., Degen J.L. Plasminogen deficiency causes severe thrombosis but is compatible with development and reproduction. Gene Dev. 1995;9(7):794–807. doi: 10.1101/gad.9.7.794. [DOI] [PubMed] [Google Scholar]
- 54.Bugge T.H., Kombrinck K.W., Flick M.J., Daugherty C.C., Danton M.J., Degen J.L. Loss of fibrinogen rescues mice from the pleiotropic effects of plasminogen deficiency. Cell. 1996;87(4):709–719. doi: 10.1016/s0092-8674(00)81390-2. [DOI] [PubMed] [Google Scholar]
- 55.Grzegorski S.J., Hu Z., Liu Y., Yu X., Ferguson A.C., Madarati H., Friedmann A.P., Reyon D., Kim P.Y., Kretz C.A., Joung J.K., Shavit J.A. Disruption of the kringle 1 domain of prothrombin leads to late onset mortality in zebrafish. Sci. Rep. 2020;10(1):4049. doi: 10.1038/s41598-020-60840-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hovhannisyan L.P., Mkrtchyan G.M., Sukiasian S.H., Boyajyan A.S. Alterations in the complement cascade in post-traumatic stress disorder. Allergy Asthma Clin. Immunol. : official journal of the Canadian Society of Allergy and Clinical Immunology. 2010;6(1):3. doi: 10.1186/1710-1492-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Laskowski and J. M. Thurman, Chapter 14 - factor B, in: S. Barnum and T. Schein (Eds.) The Complement FactsBook (second ed.), Academic Press2018, pp. 135-146.
- 58.Tawadrous H., Maga T., Sharma J., Kupferman J., Smith R.J.H., Schoeneman M. A novel mutation in the Complement Factor B gene (CFB) and atypical hemolytic uremic syndrome. Pediatr. Nephrol. 2010;25(5):947–951. doi: 10.1007/s00467-009-1415-3. [DOI] [PubMed] [Google Scholar]
- 59.Qi M., Elion E.A. MAP kinase pathways. J. Cell Sci. 2005;118(16):3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 60.Krens S.F.G., He S., Spaink H.P., Snaar-Jagalska B.E. Characterization and expression patterns of the MAPK family in zebrafish. Gene Expr. Patterns. 2006;6(8):1019–1026. doi: 10.1016/j.modgep.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 61.Kim E.K., Choi E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 62.Shi X., Zhou B. The role of Nrf 2 and MAPK pathways in PFOS-induced oxidative stress in zebrafish embryos. Toxicol. Sci. : an official journal of the Society of Toxicology. 2010;115(2):391–400. doi: 10.1093/toxsci/kfq066. [DOI] [PubMed] [Google Scholar]
- 63.Jopling C., Suñe G., Morera C., Izpisua Belmonte J.C. p38α MAPK regulates myocardial regeneration in zebrafish. Cell Cycle. 2012;11(6):1195–1201. doi: 10.4161/cc.11.6.19637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Annerén C., Lindholm C.K., Kriz V., Welsh M. The FRK/RAK-SHB signaling cascade: a versatile signal-transduction pathway that regulates cell survival, differentiation and proliferation. Curr. Mol. Med. 2003;3(4):313–324. doi: 10.2174/1566524033479744. [DOI] [PubMed] [Google Scholar]
- 65.Xiong J., Wang X., Fan C., Yan J., Zhu J., Cai T. Hemifacial microsomia is linked to a rare homozygous variant V162I in FRK and validated in zebrafish. Oral Dis. 2023;29(8):3472–3480. doi: 10.1111/odi.14372. [DOI] [PubMed] [Google Scholar]
- 66.Barr R.K., Bogoyevitch M.A. The c-Jun N-terminal protein kinase family of mitogen-activated protein kinases (JNK MAPKs) Int. J. Biochem. Cell Biol. 2001;33(11):1047–1063. doi: 10.1016/s1357-2725(01)00093-0. [DOI] [PubMed] [Google Scholar]
- 67.Pulverer B.J., Kyriakis J.M., Avruch J., Nikolakaki E., Woodgett J.R. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991;353(6345):670–674. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- 68.Wu M., Pan C., Chen Z., Jiang L., Lei P., Yang M. Bioconcentration pattern and induced apoptosis of bisphenol A in zebrafish embryos at environmentally relevant concentrations. Environ. Sci. Pollut. Res. Int. 2017;24(7):6611–6621. doi: 10.1007/s11356-016-8351-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.