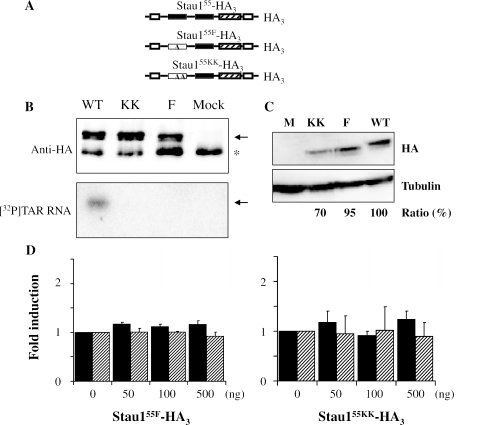
Figure 5.
The Stau155 RNA-binding activity is required for TAR-bearing RNA translational regulation. (A) Schematic representation of wild-type Stau155-HA3 and of two mutants with disrupted RNA-binding activity. Symbols are described in the legend of Figure 1. (B) HEK293T cells were transfected with plasmids coding for either Stau155-HA3, Stau155F-HA3 or Stau155KK-HA3. Tagged proteins were immunoprecipitated with an anti-HA antibody and resulting proteins were analysed by SDS–PAGE. Western blotting (upper panel) using anti-Stau1 antibody and Northwestern blotting (lower panel) using [32P]labelled TAR RNA. Arrows indicate the position of Stau1-HA3. *Non-specific IgG bands. (C) HEK293T cells were transfected with plasmids coding for either Stau155-HA3, Stau155F-HA3 or Stau155KK-HA3 and proteins were analysed by SDS–PAGE and western blotting using anti-Stau1 and anti-tubulin antibodies. Similar amounts of proteins are expressed by the three plasmids. (D) HEK293T cells were co-transfected with plasmids expressing either Rluc or TAR-Rluc transcripts and different concentrations of plasmids coding for either Stau155F-HA3 (left panel) or Stau155KK-HA3 (right panel). Resulting luciferase activity was quantified as described in Figure 2B. In the absence of Stau155-HA3, a 2-fold repression of translation of the TAR-Rluc RNA was observed as compared with translation of Rluc RNA. To facilitate comparison, fold induction of the luciferase activity in the absence of Stau155-HA3 was defined as 1. Differences are not statistically significant (n = 3). Black bars, TAR-Rluc RNA; hatched bars, Rluc RNA.