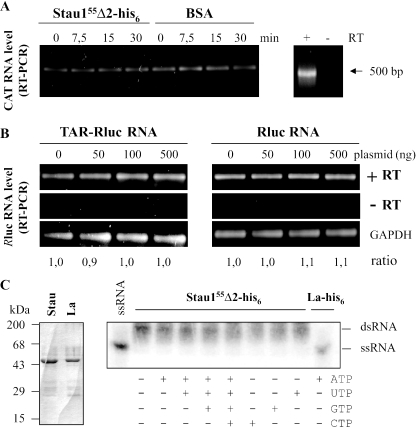
Figure 6.
Stau155 mediated translational up-regulation does not involved RNA modification. (A) TAR-CAT RNA was incubated in RRL in the presence of 400 nM of bacterially expressed and purified Stau155Δ2-his6 or BSA for increasing periods of time. TAR-CAT RNA was then reverse transcribed and PCR amplified for 14 cycles to stay in the non-saturated part of the amplification curve. Resulting DNA was resolved on agarose gel. As control, the same experiment was performed in the absence of reverse transcriptase (right panel). (B) HEK293T cells were co-transfected with plasmids expressing either Rluc or TAR-Rluc transcripts and different concentrations of a plasmid coding for Stau155-HA3. Twenty-four hours post-transfection, RNA was isolated, reverse transcribed and PCR amplified. Resulting DNA was resolved on agarose gel. As control, the same experiment was performed in the absence of reverse transcriptase (−RT). RNA coding for GAPDH was RT–PCR and used to normalize the results. (C) Bacterially expressed and column-purified Stau155Δ2-his6 (Stau) and La-his6 (La) (left panel) were incubated with [32P]labelled double-stranded RNA in the presence of different combinations of ribonucleotides (right panel). RNA was resolved on agarose gel and revealed by autoradiography. While La-his6 displayed an helicase activity, Stau155Δ2-his6 was inactive in this assay.