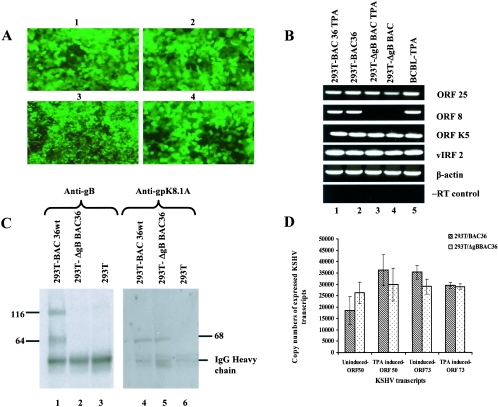
FIG. 3.
(A) GFP expression in 293T cells stably transfected with BAC36wt and ΔgBBAC36-KSHV genomes. Cells were transfected with BAC36wt and ΔgBBAC36-KSHV genomes in Lipofectamine 2000 and peptide enhancer and selected with hygromycin, and clones were established. Images 1 and 2, 293T-BAC36wt-KSHV genome-containing cells. Images 3 and 4, 293T-ΔgB BAC36-KSHV genome-containing cells. Magnification, ×40. (B) RT-PCR confirmation of the absence of gB in 293T-ΔgBBAC36 cells. Total RNAs from uninduced or TPA-induced 293T-BAC36wt and 293T-ΔgBBAC36 cells were prepared, DNase treated, and used for cDNA synthesis. cDNA (250 ng) was used in each PCR and tested for KSHV ORFs 25, 8, and K5 and vIRF2. The RNA quantity was assessed by the amplification of β-actin transcript. Absence of contaminating DNA in the samples was tested by reverse transcriptase-negative (−RT) control reactions. (C) Immunoprecipitation confirmation of the absence of gB in 293T-ΔgBBAC36-KSHV cells. Lysates from 293T-BAC36wt and 293T-ΔgBBAC36 cells induced with TPA for 96 h were immunoprecipitated with rabbit anti-gB polyclonal antibodies (lanes 1 to 3) and anti-gpK8.1A monoclonal antibodies (lanes 4 to 6). Immunoprecipitated proteins were immunoblotted with the respective antibodies and tested with alkaline phosphatase-conjugated secondary antibodies. The numbers indicate the molecular masses (in kilodaltons) of the gB and gpK8.1A proteins detected. (D) Quantitation of KSHV ORF 50 and 73 transcripts by real-time RT-PCRs. Uninduced or TPA-induced 293T-BAC36wt and 293T-ΔgBBAC36 cells were collected, and total RNA was prepared, DNase treated, and used for cDNA synthesis. DNase-treated RNA (250 ng) was subjected to real-time RT-PCR with ORF 73 and 50 gene-specific primers and Taqman probes. Known concentrations of DNase-treated in vitro-transcribed ORF 50 and ORF 73 transcripts were used in a real-time RT-PCR to construct a standard graph from which the absolute copy numbers of viral transcripts were calculated and normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcript copies used as the internal control. Each reaction was done in duplicate, and each bar represents the average ± standard deviation of three experiments.