Fig. 3.
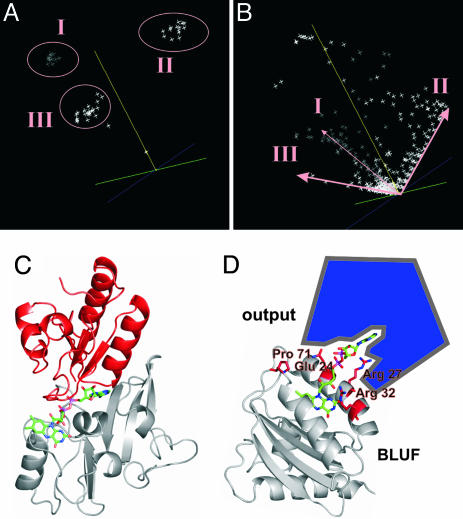
A cofactor-mediated interaction/communication between BLUF domains and output modules is suggested by the analogy with the PCMH (p-cresol-methylhydroxylase) FAD-fold family and by sequence analysis of >50 BLUF domains. (A) Sequence space representation of >50 BLUF domains allows identification of three BLUF subfamilies, I, II, and III. BlrB belongs to subfamily III. (B) Sequence space representation of the entirety of residues covered by the 50 BLUF domains analyzed. Amino acid residues specific for these families are arranged along the edges of the space, respectively. The length of a residue vector in sequence space is proportional to its degree of conservation throughout the domain family. (C) In p-cresol-methylhydroxylase (PDB ID code 1DII), FAD is bound to two distinct subdomains of the protein, shown in gray and red, that interact predominantly with the isoalloxazine or the adenine moiety of the cofactor, respectively. (D) The surface exposed adenine unit of the BLUF-FAD could function as recognition site for effector/output domains. Residues that are unique in BlrB or specific for all members of the BLUF subfamily III and that are potentially involved in the signaling process as predicted by sequence space analysis (supporting information) cluster around the entrance of the flavin pocket (red). These residues (Glu-24, Arg-27, Arg-32, and Pro-71) are predestined to serve light-dependent modulation of the BLUF domain–output domain interaction.