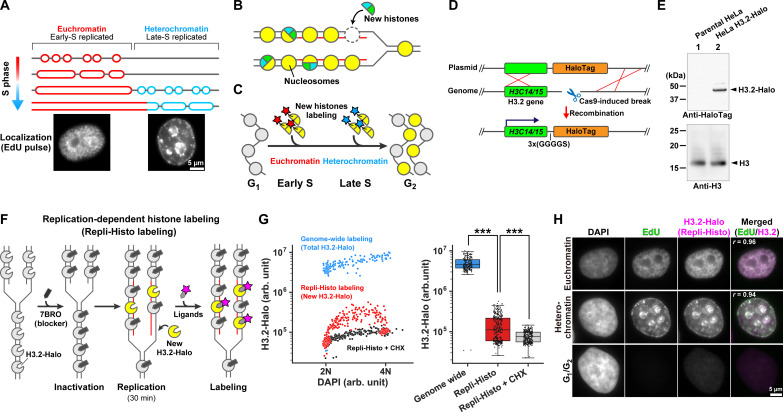
Fig. 1. Development of the Repli-Histo labeling.
(A) Eukaryotic cells replicate euchromatin in the early S phase, followed by heterochromatin replication in the late S phase. (B) Newly synthesized histones are incorporated into the replicated nucleosomes. (C) Pulse labeling of the new histones in the early and late S phases allows visualization of euchromatin and heterochromatin, respectively. (D) Schematics of Cas9-mediated HaloTag fusion with endogenous H3.2 genes (H3C14/15). (E) Western blots of H3.2-Halo in lysates of HeLa cells (lane 2) using an anti-HaloTag antibody (top) and an anti-histone H3 antibody (bottom). (F) Schematic of the replication-dependent histone labeling (Repli-Histo labeling). (G) Left: A two-dimensional plot of 4′,6-diamidino-2-phenylindole (DAPI) and tetramethylrhodamine (TMR)–labeled H3.2-Halo fluorescence in nuclei. Cyan, genome-wide H3.2-Halo labeling (n = 192 cells); red, Repli-Histo labeling (n = 326 cells); black, Repli-Histo labeling with cycloheximide (CHX) treatment (n = 316 cells). Right: The fluorescent intensity of H3.2-Halo on the left. ***P = 1.9 × 10−77 for genome-wide H3.2-Halo and Repli-Histo labeling and P = 4.62 × 10−15 for Repli-Histo labeling and Repli-Histo labeling + CHX by two-sided Mann-Whitney U test. (H) Representative HeLa cells with DAPI staining, EdU pulse labeling, and Repli-Histo labeling. The rightmost column shows the merged image with the Pearson’s correlation coefficient (green, EdU–Alexa 488; magenta, Repli-Histo labeling). Top: Euchromatin labeling; middle: heterochromatin labeling; bottom: G1-G2 cell without EdU/Repli-Histo signals.