Abstract
Mutations at position 306 of embB (embB306) have been proposed as a marker for ethambutol resistance in Mycobacterium tuberculosis; however, recent reports of embB306 mutations in ethambutol-susceptible isolates caused us to question the biological role of this mutation. We tested 1,020 clinical M. tuberculosis isolates with different drug susceptibility patterns and of different geographical origins for associations between embB306 mutations, drug resistance patterns, and major genetic group. One hundred isolates (10%) contained a mutation in embB306; however, only 55 of these mutants were ethambutol resistant. Mutations in embB306 could not be uniquely associated with any particular type of drug resistance and were found in all three major genetic groups. A striking association was observed between these mutations and resistance to any drug (P < 0.001), and the association between embB306 mutations and resistance to increasing numbers of drugs was highly significant (P < 0.001 for trend). We examined the association between embB306 mutations and IS6110 clustering (as a proxy for transmission) among all drug-resistant isolates. Mutations in embB306 were significantly associated with clustering by univariate analysis (odds ratio, 2.44; P = 0.004). In a multivariate model that also included mutations in katG315, katG463, gyrA95, and kasA269, only mutations in embB306 (odds ratio, 2.14; P = 0.008) and katG315 (odds ratio, 1.99; P = 0.015) were found to be independently associated with clustering. In conclusion, embB306 mutations do not cause classical ethambutol resistance but may predispose M. tuberculosis isolates to the development of resistance to increasing numbers of antibiotics and may increase the ability of drug-resistant isolates to be transmitted between subjects.
The antibiotic ethambutol (EMB) appears to inhibit the growth of both Mycobacterium tuberculosis and Mycobacterium smegmatis by blocking the synthesis of arabinogalactan. Arabinogalactan biosynthesis is dependent on the activity of the embABC gene cluster, which encodes the arabinotransferases that mediate the polymerization of arabinose into arabinan. Several lines of evidence suggest that EMB exerts its toxic effect on mycobacteria by inhibiting the embABC-encoded proteins (26, 27, 34, 35, 46), and mutations in embABC also appear to play a key role in the development of EMB resistance in both M. tuberculosis and M. smegmatis (34). Associations between EMB resistance and mutations in embA, embB, and embC have been reported in clinical strains of M. tuberculosis (41), and mutations in codon 306 of the embB gene (embB306) in M. tuberculosis are seen in approximately 50% of all EMB-resistant clinical isolates. In the six embB306 nucleotide polymorphisms that have been described, the wild-type methionine is changed to either isoleucine, leucine, valine, or threonine (12, 31, 33, 34, 36, 41, 45, 52). Each mutation has been associated with an 8- to 16-fold increase in the MIC of EMB (1, 33, 41, 45).
The association between embB306 mutations and EMB resistance in clinical M. tuberculosis isolates is so strong that it has been proposed as a marker for EMB resistance in diagnostic tests (13, 33, 36, 43, 52, 54). Thus, it was a surprise when Mokrousov et al. (37) first described 48 clinical M. tuberculosis isolates from Russia that were susceptible to EMB yet that had mutations in embB306. All of these EMB-susceptible embB306 mutants were resistant to at least one antibiotic. Several other small studies confirmed these findings (31, 36, 39, 48). We hypothesized that EMB might have two function: one that inhibits the growth of M. tuberculosis at concentrations above its MIC and another that prevents the development of resistance to antibiotics other than EMB at concentrations below its MIC. We postulated that the second function might be abrogated by embB306 mutations, even in the absence of overt EMB resistance. This hypothesis was consistent with data suggesting that the embABC gene cluster is responsible for the biosynthesis of several different cell wall components (10, 13, 35, 57) and prior work showing that sub-MICs of EMB are synergistic with other antibiotics (13, 20). Confirmation of this possibility might have an important impact on tuberculosis treatment strategies (6).
In this study, 1,020 clinical M. tuberculosis isolates with different drug susceptibility patterns and of different geographical origins were tested for detection of associations between embB306 (and other) mutations, drug resistance patterns, and membership in a cluster defined by restriction fragment length polymorphism (RFLP) analysis. We found that embB306 mutations were strongly associated with resistance to increasing numbers of drugs and were independently associated with membership in a cluster. These results suggest that one function of EMB is prevention of the emergence of resistance to other antibiotics and that this function is distinct from its ability to inhibit the growth of M. tuberculosis. The development of embB306 mutations may predispose M. tuberculosis to the development of resistance to multiple antibiotics and may increase the ability of these multiple-drug-resistant isolates to be transmitted between subjects.
MATERIALS AND METHODS
M. tuberculosis clinical isolates.
A total of 1,020 clinical M. tuberculosis isolates were obtained from Australia, Colombia, India, Mexico, New York City, Spain, and Texas. The 56 samples from Australia consisted of all of the isolates resistant at least to isoniazid (INH) collected in the state of Victoria at the Victorian Mycobacterium Reference Laboratory in Melbourne during 2001 and 2002 and 26 pansusceptible isolates. The samples from Colombia consisted of 142 isolates resistant to at least rifampin (RIF) and 163 pansusceptible isolates selected from the strain bank of the Mycobacterial Group, Instituto Nacional de Salud, composed of clinical M. tuberculosis isolates collected from all regions of Colombia between 1992 and 2000. The samples from India consisted of all 38 drug-resistant isolates of any type identified in the Vallabhbhai Patel Chest Institute in New Delhi between January 2001 and January 2002 plus 10 randomly selected susceptible control isolates recovered during the same period. The samples from Mexico consisted of 197 clinical isolates selected from a sample collection of the Laboratory of Clinical Microbiology, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, collected in Mexico City, Huauchinango, Puebla, and Orizaba between 1991 and 1996 (7, 16, 24). The samples from Spain consisted of all isolates collected at the General Penitentiary Hospital Microbiology Laboratory from a longitudinal study of tuberculosis performed between January 1993 and June 1994 (11). The samples from Texas consisted of all resistant isolates collected by the Texas Department of Health and the University of Texas Health Center at Tyler between May 1992 and August 1994 and 91 sensitive isolates collected in a population-based study of Tarrant County between May 1992 and December 1996 (55, 56) that were submitted to the mycobacteriology research laboratory at Central Arkansas Veterans Healthcare System for genotype analysis. Samples from New York City consisted of a random selection of all isolates collected at the Montefiore Medical Center, Bronx, between 1989 and 1996 (2).
Antibiotic susceptibility testing and cluster analysis.
All isolates included in this study were subjected to susceptibility testing and DNA fingerprinting as described below. Each center or laboratory tested the isolates for their susceptibilities to at least INH, RIF, streptomycin (STR), and EMB by the agar proportion method (25) (Colombia, India, New York, and Spain), with the BACTEC MGIT 960 system (21) (Australia), or by the radiometric BACTEC 460 method (32) (Mexico and Texas); extracted chromosomal DNA; and performed IS6110-based RFLP analysis (2, 5). MICs for selected isolates were determined with the BACTEC system (19) for isolates from Mexico and Australia or by the colorimetric assay (14) for isolates from Colombia. Isolates were considered to be clustered if they had an RFLP pattern identical to that of another isolate collected from the same study site over the study period. Isolates with low band numbers were confirmed to be identical by secondary methods (variable numbers of tandem repeats and spoligotyping) (15, 22). A description of the isolates included in the study is shown in Table 1.
TABLE 1.
Clinical M. tuberculosis isolates included in the study
| Geographic origin | No. (%) of isolates
|
No. (%) of isolates resistant to the following no. of antibiotics:
|
No. (%) of isolates with the following membershipb:
|
No. (%) of embB306 mutants | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Pansusceptible | EMBra | One | Two | Three | Four | Clustered | Unique | ||
| Australia | 56 | 26 (46) | 4 (7) | 21 (38) | 6 (11) | 3 (5) | 0 | 0 | 56 (100) | 1 (2) |
| Colombia | 304 | 163 (54) | 42 (14) | 7 (2) | 31 (10) | 72 (24) | 31 (10) | 214 (70) | 90 (30) | 48 (15) |
| India | 48 | 10 (21) | 10 (21) | 13 (27) | 14 (29) | 8 (17) | 3 (6) | 12 (25) | 36 (75) | 4 (8) |
| Mexico | 197 | 73 (37) | 17 (9) | 46 (23) | 51 (26) | 15 (8) | 9 (5) | 56 (28) | 141 (72) | 24 (13) |
| New York City | 149 | 121 (82) | 11 (7) | 7 (5) | 8 (5) | 2 (1) | 10 (7) | 66 (44) | 83 (56) | 12 (8) |
| Spain | 109 | 98 (90) | 3 (3) | 1 (1) | 6 (5) | 4 (4) | 0 | 69 (64) | 39 (36) | 0 |
| Texas | 157 | 91 (58) | 21 (13) | 30 (19) | 11 (7) | 8 (5) | 17 (11) | 58 (37) | 99 (63) | 11 (7) |
| Total | 1,020 | 582 (57) | 108 (10) | 122 (12) | 130 (13) | 112 (11) | 69 (7) | 372 (44) | 480 (56) | 100 (10) |
EMBr, resistant to EMB. The EMB resistance breakpoints used were 2 μg/ml in Spain, 2.2 μg/ml in Colombia, 2.5 μg/ml in Australia and New York City, 5 μg/ml in India and Texas, and 7.5 μg/ml in Mexico.
Isolates clustered by IS6110-based RFLP analysis. One pansusceptible isolate from Spain was not typed by RFLP analysis.
Mutation detection.
Mutations in katG315, katG463, gyrA95, and kasA269 were detected by hairpin primer (HP) assays, as described previously (17), with the primers listed in Table 2. Mutations in katG315 were confirmed by a previously described molecular beacon assay (38). Mutations in embB306 were screened by a set of four molecular beacons in which the molecular beacon BembB306 fluoresced brightly in real-time PCRs containing wild-type embB306 sequences while molecular beacons BembBM306L-c, BembBM306I-c, and BembBM306I-a detected common mutant embB306 alleles (Table 2). Samples that were negative by all molecular beacon assays were directly sequenced to identify the mutation present. A subset of samples that were positive for each mutant molecular beacon was also sequenced to confirm the accuracy of each assay. All molecular beacon assays were performed at 60°C, following the recommendations described previously (49, 50).
TABLE 2.
Molecular beacons, sequencing primers, and HPs used in this study
| Molecular beacon or primer name | Sequencea | Remarks | Amplicon length (bp) |
|---|---|---|---|
| BembB306 | FAMb-cgcgacatcctgggcatggccgtcgcg-Dc | Wild-type (ATG)dmolecular beacon | |
| BembBM306L-c | TETe-cgcgacatcctgggcCtggccgtcgcg-D | Mutant (CTG)fmolecular beacon | |
| BembBM306I-c | FAM-cgcgacatcctgggcatCgccgtcgcg-D | Mutant (ATC)gmolecular beacon | |
| BembBM306I-a | FAM-cgcgacatcctgggcatAgccgtcgcg-D | Mutant (ATA)gmolecular beacon | |
| FB2embB306 | cgaattcgtcggacgacg | PCR primers for beacon assay | 65 |
| RB2embB306 | catgtagccggcgtggtc | ||
| FSeqembB306 | accgacgccgtggtgata | Sequencing primers | 129 |
| RBembB306 | gcggaaatagttggacatgtagc | ||
| RHPgyrA95 | gcctgggccatCcgcaccaggc | Constant primer gyrAF3, cgagaccatgggcaactaccacc | 69 |
| RHPgyrA95mut | ccctgggccatCcgcaccaggG | ||
| FHPkatG463 | cggatctagcctttagagccagatccg | Constant primer RkatG463, gagacagtcaatcccgatgc | 41 |
| FHPkatGR463L | aggatctgagcctttagagccagatccT | ||
| FHP-A-katG315 | tggtgatcggtaaggacgAgatcacca | Constant primer RkatG321, aaactgttgtcccatttcgtcgg | 70 |
| FHP-A-katGS315R | gggtgatcggtaaggacgAgatcaccC | ||
| FHP-katG315h | ctggtgatcggtaaggacgAgatcaccag | Constant primer RkatG321, aaactgttgtcccatttcgtcgg | 71 |
| FHP-katGS315Lh | agggtgatcggtaaggacgAgatcaccCT | ||
| RHPkatG315 | gcggccatacgTcctcgatgccgc | Constant primer FkatG315, ccggtaaggacgcgatcac | 40 |
| RHPkatGS315I | tcggccatacgTcctcgatgccgA | ||
| RHPkatGS315N | acggccatacgTcctcgatgccgT | ||
| RHPkatGS315T | ccggccatacgTcctcgatgccgG | ||
| RHPkatGS315Tdouble | caggcacatacgTcctcgatgccTG | ||
| FHPkasA269 | cggcacgattCctgggtgccg | Constant primer RkasA269, aaaggcgtccgaggtgatac | 36 |
| RHPKasAG269S | tggcacgattCctgggtgccA |
Underlined sequences indicate the stem of the molecular beacon or the tail of the HP; residues shown in capital letters correspond to either the mutated residue, a secondary mutation inserted into the oligonucleotide, or the intended SNP.
FAM, 6-carboxyfluorescein.
D, [4-(4′-dimethylaminophenylazo)benzoic acid]succimidyl ester (DABCYL).
Encodes a methionine.
TET, tetrachlorofluorescein.
Encodes a leucine.
Encodes an isoleucine.
Assays for other katG alleles are described previously (17).
DNA sequencing.
The embB306 position was amplified by PCR and sequenced with primers FSeqembB306 and RBembB306 (Table 2). DNA sequencing was performed with a Dye Terminator kit (Applied Biosystems), and the reactions were analyzed with an ABI 3100 genetic analyzer.
Statistical analysis.
Chi-square tests for trend with and without adjustment for country of origin were performed to test whether the proportion of mutant embB306 isolates increased with the number of drugs to which an isolate was resistant. Pearson chi-square tests were used to test for an association between the embB306 genotype and clustering. We employed an overall multifactor logistic regression model to evaluate whether independent associations existed between clustering and the chosen set of single-nucleotide polymorphisms (SNPs). All mutations were initially entered into the overall model, and individual SNPs found to be insignificant were sequentially removed from the model. The best-fitting model was obtained for resistant isolates obtained from studies that took place over 2 years and resistant cases in general. SAS version 8.02 was used for all statistical analyses.
RESULTS
Associations between embB306 mutations and drug resistance.
A total of 1,020 clinical isolates of M. tuberculosis were evaluated by the embB306 molecular beacon assays and DNA sequencing. One hundred isolates (10%) contained a mutation in embB306. Forty-six of the embB306 mutants were susceptible to EMB, while 54 were EMB resistant (Table 3). Mutants were present in samples from all countries except Spain at frequencies between 2% and 15%; samples from Spain did not have any embB306 mutants (Table 1). The susceptibilities to EMB of 104 isolates from Australia, Colombia, India, Mexico, Spain, and Texas were retested. Twenty-eight of these isolates were embB306 mutants; however, the investigators responsible for retesting EMB susceptibility were blinded to the earlier results. Only five isolates (three with embB306 mutations) were found to have discrepant susceptibilities, with resistant isolates falsely classified as susceptible. The results for all samples with discrepant results were reconfirmed at least one more time prior to the final analysis. We also determined the EMB MICs for 55 isolates from Australia, Colombia, and Mexico, including 12 embB306 mutants. The EMB MICs ranged from <1 to >32 μg/ml, and no association was observed between the embB306 genotype and the MIC (Table 4). Notably, the isolate with the lowest EMB MIC was found to have an embB306 mutation.
TABLE 3.
Distribution of embB306 mutations in clinical M. tuberculosis isolates used in this study
| Drug resistancea | No. of isolates with the following genotype at embB306:
|
No. of embB306 mutants/no. of wild-type isolates (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Wild-type ATGb | Mutant
|
|||||||
| CTGc | TTGc,d | GTGe | ATCf | ATAf | ATTf | |||
| Pansusceptible | 582 | 0/582 (0) | ||||||
| INH | 93 | 1 | 1 | 2 | 4/97 (4) | |||
| RIF | 9 | 0/9 (0) | ||||||
| STR | 18 | 0/18 (0) | ||||||
| EMB | 1 | 0/1 (0) | ||||||
| INH + RIF | 76 | 2 | 5 | 14 | 21/97 (22) | |||
| INH + STR | 23 | 2 | 2/25 (8) | |||||
| INH + EMB | 1 | 1 | 1/2 (50) | |||||
| RIF + STR | 1 | 0/1 (0) | ||||||
| RIF + EMB | 0 | 1 | 1/1 (100) | |||||
| STR + EMB | 4 | 0/4 (0) | ||||||
| INH + RIF + STR | 64 | 11 | 1 | 6 | 1 | 19/83 (23) | ||
| INH + RIF + EMB | 15 | 2 | 2 | 2 | 2 | 1 | 9/24 (37) | |
| RIF + STR + EMB | 2 | 0/2 (0) | ||||||
| INH + STR + EMB | 1 | 1 | 1 | 2/3 (67) | ||||
| INH + RIF + STR + EMB | 29 | 3 | 20 | 2 | 13 | 3 | 41/70 (59) | |
| Resistant to one or more antibiotics | 338 | 6 | 1 | 35 | 12 | 41 | 5 | 100/438 (23) |
| Total | 920 | 6 | 1 | 12 | 35 | 41 | 5 | 100/1,020 (10) |
The antibiotics tested were INH, RIF, STR and EMB.
Encodes a methionine.
Encodes a leucine.
Novel mutation.
Encodes a valine.
Encodes an isoleucine.
TABLE 4.
MICs of EMB for selected wild-type and mutant isolates with mutations at position embB306
| Isolate | EMB susceptibility | EMB MIC (μg/ml)a | Genotype at embB306 |
|---|---|---|---|
| Mexico 111 | Susceptible | <1 | Mutant (ATA) |
| Colombia 717 | Susceptible | 1 | Wild type |
| Colombia 125 | Susceptible | 2 | Wild type |
| Australia 2558 | Susceptible | 2.5 | Mutant (ATC) |
| H37Rv | Susceptible | 2.5 | Wild type |
| Colombia 1311 | Susceptible | 4 | Wild type |
| Colombia 1314 | Susceptible | 4 | Wild type |
| Mexico 5306 | Susceptible | 6 | Mutant (ATA) |
| Mexico 5523 | Susceptible | 6 | Mutant (ATA) |
| Mexico 5041 | Moderately resistant | 8b | Mutant (CTG) |
| Mexico 12563 | Moderately resistant | 8 | Mutant (ATA) |
| Colombia 1194 | Moderately resistant | 8b | Wild type |
| Colombia 1299 | Moderately resistant | 8b | Wild type |
| Mexico 5036 | Resistant | 10 | Wild type |
| Mexico 30787 | Resistant | 12 | Wild type |
| Colombia 003 | Resistant | 16 | Wild type |
| Colombia 444 | Resistant | 16 | Wild type |
| Colombia 959 | Resistant | 16 | Wild type |
| Colombia 828 | Resistant | 16 | Mutant (ATC) |
| Mexico 5309 | Resistant | >16 | Mutant (CTG) |
| Mexico 30167 | Resistant | >16 | Mutant (ATC) |
| Mexico 16703 | Resistant | >16 | Wild type |
| Colombia 227 | Resistant | >32 | Mutant (GTG) |
| Colombia 1125 | Resistant | >32 | Mutant (GTG) |
| Colombia 1231 | Resistant | >32 | Mutant (ATT) |
The breakpoint value is 7.5 μg/ml.
The EMB susceptibilities of these isolates were reconfirmed twice by the agar proportion method, but the MIC classifies them as moderately resistant.
Mutations in embB306 can result in several different amino acid substitutions (12, 31, 33, 34, 36, 41, 45, 52). We examined each amino acid change separately to determine if any particular substitution had a unique association with EMB resistance. However, no such associations were observed (Tables 3 and 4). A striking association was observed between embB306 mutations and drug resistance in general. Thus, none of the 582 pansusceptible isolates contained an embB306 mutation, while 100 of 438 (23%) of the isolates that were resistant to at least one antibiotic contained a mutation at this locus (P < 0.001 for association with broad drug resistance). This association may explain why the Spanish samples did not include any embB306 mutants, as only 11 of 109 (10%) of the Spanish isolates were resistant to any drug.
The overall drug resistance patterns of the embB306 mutants were examined in order to determine whether embB306 mutations could be responsible for resistance to any antibiotic other than EMB. Nine of the 100 embB306 mutants were susceptible to RIF, 36 were susceptible to STR, and 1 was susceptible to INH (Table 3). We considered the possibility that a mistake had been made in the analysis of the single INH-susceptible embB306 mutant; it was retested twice to confirm its antibiotic susceptibility, and the presence of the embB306 mutation was also reconfirmed by direct DNA sequencing. We also determined that each of the four INH-monoresistant embB306 mutants contained other mutations that could account for their resistance to INH using assays described previously (38) (Table 5), and their patterns of susceptibility to all antibiotics were reconfirmed. These results strongly indicate that embB306 mutations are not one of the causes of INH resistance. Thus, mutations in embB306 could not be uniquely associated with any particular type of drug resistance.
TABLE 5.
INH resistance-associated mutations detected in the INH monoresistant embB306 mutants
| Isolate | Mutation at embB306 | INH resistance-associated mutations | Major genetic group |
|---|---|---|---|
| Australia 2558 | M306I (ATG→ATC) | inhA promoter (−17 G→T) | I |
| inhAI194T (ATC→ACC) | |||
| ahpC promoter (−46 G→A) | |||
| Mexico 111 | M306I (ATG→ATA) | katGS315T (AGC→ACC) | II |
| Mexico 5041 | M306L (ATG→CTC) | inhA promoter (−15 C→T) | II |
| inhAS94A (TCG→GCG) | |||
| Mexico 5306 | M306I (ATG→ATA) | katGS315T (AGC→ACC) | II |
Genotype at position embB306 after in vitro selection for EMB resistance.
Approximately 50% of EMB-resistant clinical isolates have embB306 mutations (12, 31, 33, 36, 37, 43, 52). One would expect to find a similar proportion of embB306 mutants during in vitro selection for EMB resistance, if this mutation truly conferred resistance to EMB. Thirty-seven EMB-resistant colonies were isolated after approximately 4 × 107 CFU of strain H37Rv was plated onto Middlebrook 7H10 medium containing 20 μg/ml of EMB. All 37 colonies were found to have the wild-type sequence at the embB306 codon. We also tested whether lower concentrations of EMB could select for embB306 mutations in isolates that were grown at the MIC of EMB. H37Rv was plated on medium containing 2.5 μg/ml of EMB. All of the 55 colonies isolated under these conditions were EMB susceptible; none contained an embB306 mutation. We also considered the possibility that embB306 mutations did indeed confer EMB resistance but that the resistant phenotype was being suppressed by another mutation in the embB gene. The complete embB gene of two EMB-susceptible clinical strains (Australia 2558 and Mexico 5041) with embB306 mutations was sequenced to test this hypothesis. Both EMB-susceptible strains had wild-type embB sequences except at embB306.
Phylogenetic studies.
We considered the possibility that the association between the embB306 mutations and drug resistance was not due to a change in the biological activity of the embB gene. It was possible that embB306 mutations were simply phylogenetic markers for a related family of isolates that were predisposed to the development of drug resistance through a mechanism unrelated to the embB gene activity. This hypothesis would have been consistent with reports that the Beijing family of M. tuberculosis is predisposed to the development of multidrug resistance (4, 28, 47). The embB306 mutants were examined for membership in the three major genetic groups of M. tuberculosis (44). Mutants were found in all three major genetic groups: 22 (22%) in group I, 66 (66%) in group II, and 12 (12%) in group III. These findings suggest a biological rather than a phylogenetic role for embB306 mutations (3).
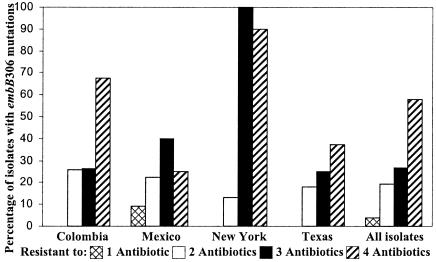
Association between embB306 mutants and resistance to increasing numbers of antibiotics.
Based on the results described above, we questioned whether embB306 mutations could confer a general predisposition to the development of resistance to any antibiotic. We reasoned that factors with the ability to nonspecifically potentiate drug resistance should be observed at increasing frequencies in isolates with resistance to an increasing number of drugs. Analysis of all 438 drug-resistant isolates revealed a strong association between the presence of the embB306 mutations and resistance to increasing number of antibiotics (P < 0.001 for trend) (Fig. 1). This association remained significant even after geographic region was controlled for as a confounder (P < 0.001). Furthermore, this trend was essentially unchanged when the effect of the IS6110-defined cluster size was removed by examining each drug resistance pattern separately and counting each cluster as a single isolate (for this analysis, a cluster that contained isolates with both wild-type and embB306 mutant sequences was counted twice but was assigned to both the mutant and the wild-type categories) (data not shown).
FIG. 1.
Proportion of drug-resistant M. tuberculosis isolates with embB306 mutations. The proportion of embB306 mutants is shown by geographical origin and the number of antibiotics to which each isolate was resistant. Only geographical sites in which at least 10 embB306 mutant isolates were identified are shown. The “all isolates” category corresponds to all isolates included in the study.
Association between embB306 mutants and clustering as a marker for transmissibility.
Several recent studies have suggested that multidrug-resistant M. tuberculosis strains may be transmitted less frequently than pansusceptible strains (9, 16). The novel association between embB306 mutations and resistance to increasing number of drugs led us to question whether this mutation could identify a subgroup of drug-resistant isolates with a heightened propensity for transmission. The clusters identified by IS6110-based RFLP analysis have become accepted as a proxy for measurements of transmission and accelerated disease progression (3, 16, 51). We found an association between embB306 mutations and presence in a cluster using a sample that consisted of all drug-resistant study isolates. This association was significant for all types of drug resistance as well as for isolates that were resistant to at least two antibiotics (Table 6). Similar results were obtained when this analysis was repeated after all isolates in each cluster were counted as a single isolate to eliminate the effect of cluster size (Table 6). This analysis was also repeated after removal of the Colombian and Australian “outlier” populations, which had the highest and lowest proportions of clustered isolates, respectively. Again, the same associations were found (data not shown). Some of the populations in this study were based on isolates collected over a relatively short period of time. Time is a known confounder in cluster analysis; therefore, the analysis was repeated a fourth time by limiting the samples to those from studies that had collected isolates over more than 2 years (which included only isolates from Colombia, Mexico, New York City, and Texas). The association between embB306 mutations and clustering remained significant in this final analysis (Table 6).
TABLE 6.
Univariate analysis showing association between embB306 mutations and clustering
| No. of antibiotics to which the isolate was resistant | embB306 genotype | All studies
|
Studies occurring over >2 yr
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. (%) of isolates in a cluster | No. (%) of unique isolates | P (χ2) | Odds ratio | No. (%) of isolates in a cluster | No. (%) of unique isolates | P (χ2) | Odds ratio | ||
| All isolates | |||||||||
| Two or more | Mutant | 65 (68) | 31 (32) | 0.017 | 1.84 | 64 (70) | 28 (30) | 0.092 | 1.58 |
| WTb | 115 (53) | 101 (47) | 104 (59) | 72 (41) | |||||
| One or more | Mutant | 67 (67) | 33 (33) | <0.001 | 2.44 | 66 (69) | 29 (31) | 0.004 | 2.06 |
| WT | 153 (45) | 184 (55) | 136 (53) | 123 (47) | |||||
| One isolate per clustera | |||||||||
| Two or more | Mutant | 22 (42) | 31 (58) | 0.034 | 2.05 | 21 (43) | 28 (57) | 0.085 | 1.86 |
| WT | 35 (26) | 101 (74) | 29 (29) | 72 (71) | |||||
| One or more | Mutant | 22 (40) | 33 (60) | 0.008 | 2.27 | 21 (42) | 29 (58) | 0.046 | 1.94 |
| WT | 54 (23) | 184 (77) | 46 (27) | 123 (73) | |||||
Each cluster was counted as a single isolate. In cases where a cluster contained isolates with both wild-type and embB306 mutant sequences, the cluster was counted twice but was assigned to both the mutant and the wild-type categories.
WT, wild type.
Multivariate analysis.
The association between embB306 mutations and clustering that we observed could have been biased by our failure to include other mutations potentially associated with clustering in our analysis. Hidden biases in the study population or biases due to sampling could also have influenced our results. We tested all drug-resistant study isolates for mutations in katG315, katG463, gyrA95, and kasA269 to control for these possibilities. Previous studies have suggested that katG315 mutations are associated with clustering and drug-resistant tuberculosis (40, 53). This mutation was chosen as the best approximation of a positive control. As negative controls we used mutations at katG463, gyrA95, and kasA269, which are known to be neutral phylogenetic markers (3, 38, 42, 44). Like embB306 mutations, these four mutations occur in genes that are involved in antibiotic resistance. Univariate analysis initially suggested that each of these mutations was associated with clustering (data not shown). However, when these SNPs were placed in a multifactor logistic regression model, only mutations at embB306 and katG315 showed an independent significant association with clustering (Table 7). In the case of katG463, we observed a significant association with unique rather than clustered isolates when all isolates were included in the analysis, but this significance was lost when we limited our analysis to study sites that provided isolates collected for more than 2 years (Table 7).
TABLE 7.
Multivariate regression model of association between various mutations and clustering
| Study | Position | Odds ratio | 95% CIa | P value |
|---|---|---|---|---|
| All studies | embB306 | 2.41 | 1.43-4.07 | 0.001b |
| katG315 | 1.74 | 1.07-2.83 | 0.027b | |
| katG463 | 0.50 | 0.29-0.86 | 0.013 | |
| kasA269 | 2.28 | 0.77-6.77 | 0.138 | |
| gyrA95 | 1.15 | 0.56-2.38 | 0.696 | |
| Studies occurring | embB306 | 2.14 | 1.22-3.76 | 0.008b |
| over >2 yr | katG315 | 1.99 | 1.15-3.44 | 0.015b |
| katG463 | 1.01 | 0.50-2.02 | 0.981 | |
| kasA269 | 2.16 | 0.72-6.47 | 0.167 | |
| gyrA95 | 1.22 | 0.58-2.57 | 0.595 |
CI, confidence interval.
Significant association with clustering.
DISCUSSION
Our results strongly suggest that embB306 mutations do not cause EMB resistance in M. tuberculosis. Rather, we found that embB306 mutations may predispose M. tuberculosis to become resistant to any antibiotic and to become multidrug resistant. A careful search for alternate explanations and hidden biases is necessary. Some of the results of this study could be due to errors in EMB susceptibility testing, which can occasionally be unreliable (17-19, 29, 30). However, the EMB-susceptible embB306 mutants were detected in six of the seven laboratories in this study, and 46 of the embB306 mutants were EMB susceptible. An error in EMB susceptibility testing of this magnitude is highly unlikely, given that our discrepancy rate is similar to that described for other reference laboratories (29, 30). We observed embB306 mutations in 50% (54 of 108) of the EMB-resistant isolates, a percentage similar to those described in previous reports (12, 31, 33, 36, 37, 43, 52). Finally, measurements of EMB MICs demonstrated that embB306 mutations could be present in highly EMB-susceptible isolates. It should also be noted that potential errors in EMB susceptibility testing would not have affected our discovery of an association between embB306 mutations and clustering, as this analysis was performed only with isolates known to be antibiotic resistant.
One might question why mutations in embB306 have been associated with EMB resistance in many prior studies. We suggest that those studies did not include the proper EMB-susceptible control isolates in their analyses. The controls in the prior studies predominantly consisted of pansusceptible M. tuberculosis isolates, while our study found embB306 mutations only among the EMB-susceptible isolates that were resistant to other antibiotics. Thus, it is easy to see how previous investigations could have erroneously concluded that embB306 mutations were associated with EMB resistance. Our results are supported by those of other studies that used controls similar to ours (31, 37, 39, 48). The biological evidence supporting a role for embB306 mutations in EMB resistance is also weak. Overexpression of embB or replacement of wild-type embB with an I289M mutation (which corresponds to embB303 in M. tuberculosis) has been shown to cause EMB resistance in M. smegmatis (34). However, to our knowledge, EMB resistance has never been documented in M. tuberculosis either by overexpressing embB or by transferring a single copy of the embB gene containing any of the mutated embB306 alleles. In the current study, we also show that M. tuberculosis isolates selected for EMB resistance in vitro do not appear to acquire mutations in embB306.
We observed that embB306 mutations were present only in isolates that were resistant to one or more drugs. One possible explanation for this observation is that embB306 mutations increase the likelihood that any type of drug resistance will develop but do not otherwise increase bacterial fitness. This hypothesis predicts that embB306 mutations will rarely, if ever, be detected in pansusceptible isolates. However, once a rare embB306 mutant acquires a drug resistance mutation, it would then have a strong evolutionary advantage over wild-type cells. It has been shown that sub-MICs of EMB can increase M. tuberculosis susceptibility to hydrophobic antibiotics by increasing cell wall permeability (13, 20). It is possible that embB306 mutations render M. tuberculosis resistant to this effect of EMB while the isolate remains fully EMB “susceptible,” as measured by MIC or antibiotic breakpoint testing. Verification of this hypothesis could have important implications for the treatment of tuberculosis.
Clustering has been widely accepted as a marker for recently transmitted M. tuberculosis with rapid progression to disease (3, 8, 16, 42, 51). Thus, our results suggest that embB306 mutations could also be associated with disease transmission. This investigation was open to a number of biases, given its retrospective design and the combined use of several studies with different enrollment and selection criteria. An effort was made to reduce potential biases by limiting analysis to drug-resistant isolates and by including other mutations in a multivariate analysis with embB306. We found that both embB306 and katG315 mutations were independently associated with clustering, while the other control mutations did not have such an association. Mutations in katG315 have been associated with clustering among drug-resistant isolates in another large study (53). Our ability to confirm these results for katG315 in the current investigation further validates our study populations and suggests that the association which we found between embB306 and clustering is valid. Further confirmation of this hypothesis must await a large population-based study that includes all tuberculosis cases over an extended period.
Mutations in katG315 are known to cause INH resistance in M. tuberculosis (23), and it is postulated that the association between katG315 mutations and clustering can be explained by the fact that this mutation is less likely to attenuate bacterial virulence than other mutations that cause INH resistance (40). In contrast, we have shown that embB306 mutations are not associated with resistance to any particular antibiotic; thus, there is no obvious biological explanation for the observed association between embB306 mutations and clustering. However, the embABC genes are intimately involved in the biosynthesis of the M. tuberculosis cell wall. Mutations in embB306 could change the M. tuberculosis cell wall in unknown ways, in the presence or absence of EMB, altering virulence properties and increasing the transmission rates or disease progression.
In conclusion, we have demonstrated that embB306 mutations in M. tuberculosis are associated with broad drug resistance and clustering of drug-resistant isolates. Our results suggest as well that embB306 and/or katG315 could serve as a marker for tuberculosis cases that are at increased risk for the development and spread of drug-resistant disease. Additional studies are required to confirm these results and to determine the biological mechanism underlying these observations. These observations could lead to new treatment approaches that would ultimately make a significant contribution to tuberculosis control.
Acknowledgments
This work was supported by Public Health Service grant AI-46669 from the National Institutes of Health, grant PI020572 from the Fondo de Investigaciones Sanitarias (Spain), and grant 55000632 from the Howard Hughes Medical Institute.
We thank Peter Small for advice and assistance in the initial phases of this study. David Alland is among a group of coinvestigators who hold patents in molecular beacon technology and receives income from licenses.
REFERENCES
- 1.Alcaide, F., G. E. Pfyffer, and A. Telenti. 1997. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob. Agents Chemother. 41:2270-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbón, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anh, D. D., M. W. Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, P. F., H. el-Hajj, S. Preston-Martin, M. D. Cave, B. E. Jones, M. Otaya, J. Pogoda, and K. D. Eisenach. 1996. Transmission of tuberculosis among the urban homeless. JAMA 275:305-307. [PubMed] [Google Scholar]
- 6.Blumberg, H. M., W. J. Burman, R. E. Chaisson, C. L. Daley, S. C. Etkind, L. N. Friedman, P. Fujiwara, M. Grzemska, P. C. Hopewell, M. D. Iseman, R. M. Jasmer, V. Koppaka, R. I. Menzies, R. J. O'Brien, R. R. Reves, L. B. Reichman, P. M. Simone, J. R. Starke, and A. A. Vernon. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603-662. [DOI] [PubMed] [Google Scholar]
- 7.Bobadilla-del-Valle, M., A. Ponce-de-León, C. Arenas-Huertero, G. Vargas-Alarcon, M. Kato-Maeda, P. M. Small, P. Couary, G. M. Ruiz-Palacios, and J. Sifuentes-Osornio. 2001. rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis identified by polymerase chain reaction single-stranded conformational polymorphism. Emerg. Infect. Dis. 7:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgdorff, M. W., M. A. Behr, N. J. Nagelkerke, P. C. Hopewell, and P. M. Small. 2000. Transmission of tuberculosis in San Francisco and its association with immigration and ethnicity. Int. J. Tuberc. Lung Dis. 4:287-294. [PubMed] [Google Scholar]
- 9.Burgos, M., K. DeRiemer, P. M. Small, P. C. Hopewell, and C. L. Daley. 2003. Effect of drug resistance on the generation of secondary cases of tuberculosis. J. Infect. Dis. 188:1878-1884. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee, D., and K. H. Khoo. 1998. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8:113-120. [DOI] [PubMed] [Google Scholar]
- 11.Chaves, F., F. Dronda, M. D. Cave, M. Alonso-Sanz, A. Gonzalez-Lopez, K. D. Eisenach, A. Ortega, L. Lopez-Cubero, I. Fernandez-Martin, S. Catalan, and J. H. Bates. 1997. A longitudinal study of transmission of tuberculosis in a large prison population. Am. J. Respir. Crit. Care Med. 155:719-725. [DOI] [PubMed] [Google Scholar]
- 12.Escalante, P., S. Ramaswamy, H. Sanabria, H. Soini, X. Pan, O. Valiente-Castillo, and J. M. Musser. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber. Lung Dis. 79:111-118. [DOI] [PubMed] [Google Scholar]
- 13.Escuyer, V. E., M. A. Lety, J. B. Torrelles, K. H. Khoo, J. B. Tang, C. D. Rithner, C. Frehel, M. R. McNeil, P. J. Brennan, and D. Chatterjee. 2001. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 276:48854-48862. [DOI] [PubMed] [Google Scholar]
- 14.Franzblau, S. G., R. S. Witzig, J. C. McLaughlin, P. Torres, G. Madico, A. Hernandez, M. T. Degnan, M. B. Cook, V. K. Quenzer, R. M. Ferguson, and R. H. Gilman. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144(Pt 5):1189-1196. [DOI] [PubMed] [Google Scholar]
- 16.García-García, M. L., A. Ponce de León, M. E. Jimenez-Corona, A. Jimenez-Corona, M. Palacios-Martinez, S. Balandrano-Campos, L. Ferreyra-Reyes, L. Juarez-Sandino, J. Sifuentes-Osornio, H. Olivera-Diaz, J. L. Valdespino-Gomez, and P. M. Small. 2000. Clinical consequences and transmissibility of drug-resistant tuberculosis in southern Mexico. Arch. Intern. Med. 160:630-636. [DOI] [PubMed] [Google Scholar]
- 17.Hazbón, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazbón, M. H., N. Guarín, B. E. Ferro, A. L. Rodriguez, L. A. Labrada, R. Tovar, P. F. Riska, and W. R. Jacobs, Jr. 2003. Photographic and luminometric detection of luciferase reporter phages for drug susceptibility testing of clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:4865-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heifets, L. B., M. D. Iseman, and P. J. Lindholm-Levy. 1986. Ethambutol MICs and MBCs for Mycobacterium avium complex and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 30:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagannath, C., V. M. Reddy, and P. R. Gangadharam. 1995. Enhancement of drug susceptibility of multi-drug resistant strains of Mycobacterium tuberculosis by ethambutol and dimethyl sulphoxide. J. Antimicrob. Chemother. 35:381-390. [DOI] [PubMed] [Google Scholar]
- 21.Johansen, I. S., V. O. Thomsen, M. Marjamaki, A. Sosnovskaja, and B. Lundgren. 2004. Rapid, automated, nonradiometric susceptibility testing of Mycobacterium tuberculosis complex to four first-line antituberculous drugs used in standard short-course chemotherapy. Diagn. Microbiol. Infect. Dis. 50:103-107. [DOI] [PubMed] [Google Scholar]
- 22.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapetanaki, S. M., S. Chouchane, S. Yu, X. Zhao, R. S. Magliozzo, and J. P. Schelvis. 2005. Mycobacterium tuberculosis KatG(S315T) catalase-peroxidase retains all active site properties for proper catalytic function. Biochemistry 44:243-252. [DOI] [PubMed] [Google Scholar]
- 24.Kato-Maeda, M., J. Sifuentes-Osornio, M. Bobadilla-del-Valle, G. M. Ruiz-Palacios, and A. Ponce-de-Leon. 1999. Drug resistance among acid-fast bacilli. Lancet 353:1709. [DOI] [PubMed] [Google Scholar]
- 25.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, Atlanta, Ga.
- 26.Khoo, K. H., E. Douglas, P. Azadi, J. M. Inamine, G. S. Besra, K. Mikusova, P. J. Brennan, and D. Chatterjee. 1996. Truncated structural variants of lipoarabinomannan in ethambutol drug-resistant strains of Mycobacterium smegmatis. Inhibition of arabinan biosynthesis by ethambutol. J. Biol. Chem. 271:28682-28690. [DOI] [PubMed] [Google Scholar]
- 27.Khoo, K. H., J. B. Tang, and D. Chatterjee. 2001. Variation in mannose-capped terminal arabinan motifs of lipoarabinomannans from clinical isolates of Mycobacterium tuberculosis and Mycobacterium avium complex. J. Biol. Chem. 276:3863-3871. [DOI] [PubMed] [Google Scholar]
- 28.Kruuner, A., S. E. Hoffner, H. Sillastu, M. Danilovits, K. Levina, S. B. Svenson, S. Ghebremichael, T. Koivula, and G. Kallenius. 2001. Spread of drug-resistant pulmonary tuberculosis in Estonia. J. Clin. Microbiol. 39:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laszlo, A., M. Rahman, M. Espinal, and M. Raviglione. 2002. Quality assurance programme for drug susceptibility testing of Mycobacterium tuberculosis in the WHO/IUATLD Supranational Reference Laboratory Network: five rounds of proficiency testing, 1994-1998. Int. J. Tuberc. Lung Dis. 6:748-756. [PubMed] [Google Scholar]
- 30.Laszlo, A., M. Rahman, M. Raviglione, and F. Bustreo. 1997. Quality assurance programme for drug susceptibility testing of Mycobacterium tuberculosis in the WHO/IUATLD Supranational Laboratory Network: first round of proficiency testing. Int. J. Tuberc. Lung Dis. 1:231-238. [PubMed] [Google Scholar]
- 31.Lee, A. S., S. N. Othman, Y. M. Ho, and S. Y. Wong. 2004. Novel mutations within the embB gene in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:4447-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, C. N., and L. B. Heifets. 1987. Determination of minimal inhibitory concentrations of antituberculosis drugs by radiometric and conventional methods. Am. Rev. Respir. Dis. 136:349-352. [DOI] [PubMed] [Google Scholar]
- 33.Lee, H. Y., H. J. Myoung, H. E. Bang, G. H. Bai, S. J. Kim, J. D. Kim, and S. N. Cho. 2002. Mutations in the embB locus among Korean clinical isolates of Mycobacterium tuberculosis resistant to ethambutol. Yonsei Med. J. 43:59-64. [DOI] [PubMed] [Google Scholar]
- 34.Lety, M. A., S. Nair, P. Berche, and V. Escuyer. 1997. A single point mutation in the embB gene is responsible for resistance to ethambutol in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 41:2629-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikusova, K., R. A. Slayden, G. S. Besra, and P. J. Brennan. 1995. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob. Agents Chemother. 39:2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mokrousov, I., N. V. Bhanu, P. N. Suffys, G. V. Kadival, S. F. Yap, S. N. Cho, A. M. Jordaan, O. Narvskaya, U. B. Singh, H. M. Gomes, H. Lee, S. P. Kulkarni, K. C. Lim, B. K. Khan, D. van Soolingen, T. C. Victor, and L. M. Schouls. 2004. Multicenter evaluation of reverse line blot assay for detection of drug resistance in Mycobacterium tuberculosis clinical isolates. J. Microbiol. Methods 57:323-335. [DOI] [PubMed] [Google Scholar]
- 37.Mokrousov, I., T. Otten, B. Vyshnevskiy, and O. Narvskaya. 2002. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from northwestern Russia: implications for genotypic resistance testing. J. Clin. Microbiol. 40:3810-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 39.Post, F. A., P. A. Willcox, B. Mathema, L. M. Steyn, K. Shean, S. V. Ramaswamy, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and G. Kaplan. 2004. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J. Infect. Dis. 190:99-106. [DOI] [PubMed] [Google Scholar]
- 40.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramaswamy, S. V., A. G. Amin, S. Goksel, C. E. Stager, S. J. Dou, H. El Sahly, S. L. Moghazeh, B. N. Kreiswirth, and J. M. Musser. 2000. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:326-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee, J. T., M. M. Tanaka, M. A. Behr, C. B. Agasino, E. A. Paz, P. C. Hopewell, and P. M. Small. 2000. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int. J. Tuberc. Lung Dis. 4:1111-1119. [PubMed] [Google Scholar]
- 43.Rinder, H., K. T. Mieskes, E. Tortoli, E. Richter, M. Casal, M. Vaquero, E. Cambau, K. Feldmann, and T. Loscher. 2001. Detection of embB codon 306 mutations in ethambutol resistant Mycobacterium tuberculosis directly from sputum samples: a low-cost, rapid approach. Mol. Cell. Probes 15:37-42. [DOI] [PubMed] [Google Scholar]
- 44.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41:1677-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telenti, A., W. J. Philipp, S. Sreevatsan, C. Bernasconi, K. E. Stockbauer, B. Wieles, J. M. Musser, and W. R. Jacobs, Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3:567-570. [DOI] [PubMed] [Google Scholar]
- 47.Toungoussova, O. S., P. Sandven, A. O. Mariandyshev, N. I. Nizovtseva, G. Bjune, and D. A. Caugant. 2002. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J. Clin. Microbiol. 40:1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tracevska, T., I. Jansone, A. Nodieva, O. Marga, G. Skenders, and V. Baumanis. 2004. Characterisation of rpsL, rrs and embB mutations associated with streptomycin and ethambutol resistance in Mycobacterium tuberculosis. Res. Microbiol. 155:830-834. [DOI] [PubMed] [Google Scholar]
- 49.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 50.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 51.van der Spuy, G. D., R. M. Warren, M. Richardson, N. Beyers, M. A. Behr, and P. D. van Helden. 2003. Use of genetic distance as a measure of ongoing transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in The Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 54.Wada, T., S. Maeda, A. Tamaru, S. Imai, A. Hase, and K. Kobayashi. 2004. Dual-probe assay for rapid detection of drug-resistant Mycobacterium tuberculosis by real-time PCR. J. Clin. Microbiol. 42:5277-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weis, S. E., J. M. Pogoda, Z. Yang, M. D. Cave, C. Wallace, M. Kelley, and P. F. Barnes. 2002. Transmission dynamics of tuberculosis in Tarrant County, Texas. Am. J. Respir. Crit. Care Med. 166:36-42. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, R. W., Z. Yang, M. Kelley, M. D. Cave, J. M. Pogoda, R. J. Wallace, Jr., J. P. Cegielski, D. F. Dunbar, D. Bergmire-Sweat, L. B. Elliott, and P. F. Barnes. 1999. Evidence from molecular fingerprinting of limited spread of drug-resistant tuberculosis in Texas. J. Clin. Microbiol. 37:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, N., J. B. Torrelles, M. R. McNeil, V. E. Escuyer, K. H. Khoo, P. J. Brennan, and D. Chatterjee. 2003. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 50:69-76. [DOI] [PubMed] [Google Scholar]