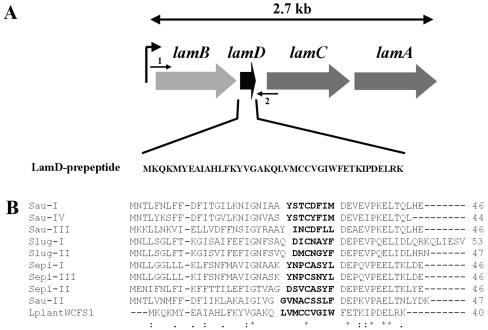
FIG. 1.
(A) Schematic representation of the gene organization of the lamBDCA operon, as deduced from the Lactobacillus plantarum WCFS1 genome project (http://www.cmbi.kun.nl/lactobacillus). The position and direction of NICE cloning primer LamBD-NICE-NcoI (arrow 1) and NICE cloning primer LamBD-NICE-KpnI (arrow 2) are shown. (B) Alignment of the L. plantarum WCFS1 LamD prepeptide with staphylococcal AgrD prepeptide sequences. The numbers of amino acids in the prepeptides are shown to the right of the sequences. Sau-I to -IV, S. aureus pherotypes I to IV; Slug-I and -II, Staphylococcus lugdunensis pherotypes I and II; Sepi-I to -III, S. epidermidis pherotypes I to III; LplantWCFS1, L. plantarum WCFS1. Amino acids of determined or predicted AIP sequences are shown in bold type. Asterisks indicate conserved amino acids in all sequences, colons indicate conserved amino acid substitutions, and dots indicate semiconserved amino acid substitutions according to physicochemical criteria. Gaps introduced to maximize alignment are indicated by dashes.