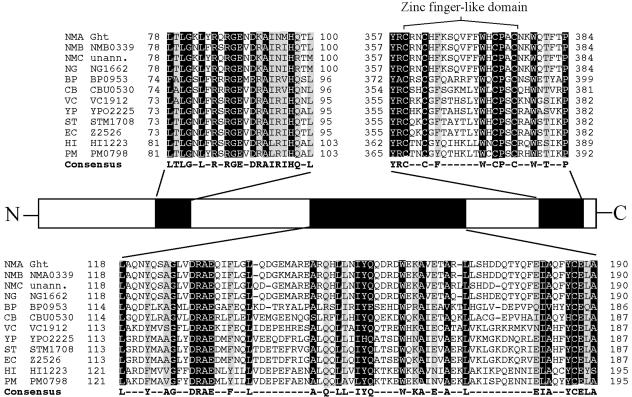
FIG. 4.
Amino acid sequence comparison of the predicted N. meningitidis group A Ght protein with homologues from 12 human pathogens. Amino acid sequences were aligned by the ClustalW method using MegAlign in the Lasergene software suite (DNAstar). Residues conserved in 11 to 12 species are highlighted in black with white writing; residues conserved in 9 to 10 species are highlighted in gray with black writing. The consensus sequence of 9 to 12 matches is found below the alignment. NMA, Neisseria meningitidis group A Z2491; NMB, Neisseria meningitidis group B MC58; NMC, Neisseria meningitidis group C FAM18; NG, Neisseria gonorrhoeae FA1090; BP, Bordetella pertussis Tohama I; CB, Coxiella burnetii RSA 493; VC, Vibrio cholerae O1 biovar El Tor N16961; YP, Yersinia pestis CO92; ST, Salmonella enterica serovar Typhimurium LT2 SGSC1412; EC, Escherichia coli O157:H7 EDL933; HI, Haemophilus influenzae RD KW20; PM, Pasteurella multocida PM70; unann., unannotated genome.