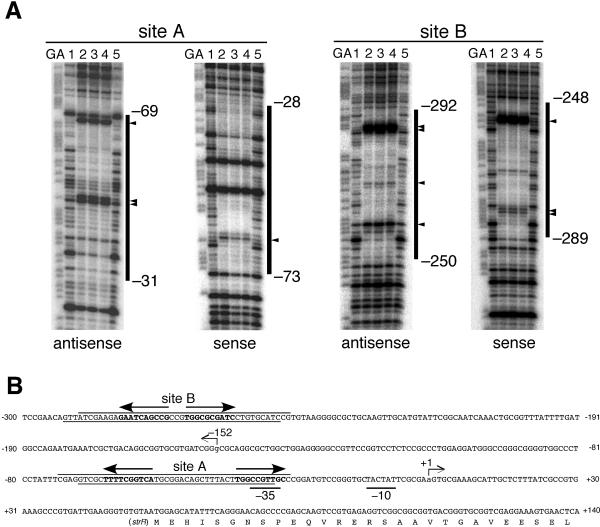
FIG. 2.
DNase I footprinting for determination of the two AdpA-binding sites upstream of strR. (A) DNase I footprinting assays were performed on the antisense and sense strands of sites A and B. The amounts of AdpA-H used in lanes 1 to 5 were 0, 0.6, 1.2, 2.4, and 0 μg, respectively. The DNase I digests were run with the same probes that were chemically cleaved (GA lanes). The nucleotide sequences protected from DNase I digestion, together with the nucleotide positions of their boundaries, are indicated. The nucleotides that are hypersensitive to DNase I are indicated by triangles. (B) Nucleotide sequence of the strR promoter and AdpA-binding sites. The arrow at position +1 indicates the transcriptional start point of strR. The arrow at position −152 indicates the transcriptional start point of strD. The DNA sequences protected from DNase I digestion are also shown, together with the AdpA-binding sequences (in boldface letters and with arrows) at sites A and B. The arrows indicate the AdpA-binding sequences (5′-TGGCSNGWWY-3′) that go from the 5′ to the 3′ side.