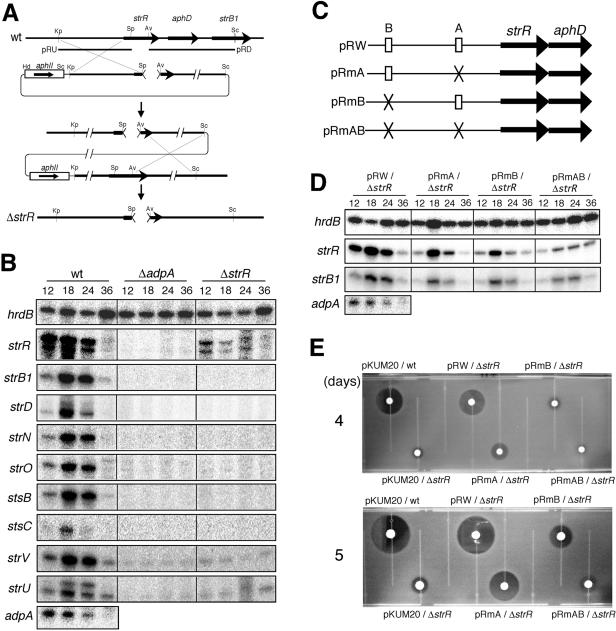
FIG. 4.
Requirement of both site A and site B for full activation of strR by AdpA. (A) Schematic representation of the procedure used for construction of the ΔstrR mutant. The restriction enzymes are abbreviated as follows: Av, AviII; Kp, KpnI; Pv, PvuII; Sc, SacI; Sp, SphI. (B) Low-resolution S1 nuclease mapping of strR and other promoters within the streptomycin biosynthetic gene cluster (strB1, strD, strN, strO, stsB, stsC, strV, and strU) in S. griseus ΔadpA and ΔstrR mutants. RNA was prepared from cells grown on Bennett agar medium at 28°C for the indicated number of hours. (C) Schematic representation of the plasmids used for promoter assays. (D) Low-resolution S1 mapping of strR and strB1 in the ΔstrR mutant harboring pRW, pRmA, pRmB, or pRmAB. (E) Streptomycin production by the ΔstrR mutant harboring pRW, pRmA, pRmB, or pRmAB. As controls, the wild-type (wt) strain harboring the vector pKUM20 and the ΔstrR mutant harboring pKUM20 were also assayed. These strains were grown on Bennett agar medium at 28°C for 4 or 5 days, and soft agar containing spores of B. subtilis was overlaid. The plates were incubated at 37°C overnight, and streptomycin production was detected by inhibition of the growth of the indicator.