Abstract
Inactivation of the lambdoid phage repressor protein is necessary to induce lytic growth of a lambdoid prophage. Activated RecA, the mediator of the host SOS response to DNA damage, causes inactivation of the repressor by stimulating the repressor's nascent autocleavage activity. The repressor of bacteriophage lambda and its homolog, LexA, preferentially undergo RecA-stimulated autocleavage as free monomers, which requires that each monomer mediates its own (intramolecular) cleavage. The cI repressor of bacteriophage 434 preferentially undergoes autocleavage as a dimer specifically bound to DNA, opening the possibility that one 434 repressor subunit may catalyze proteolysis of its partner subunit (intermolecular cleavage) in the DNA-bound dimer. Here, we first identified and mutagenized the residues at the cleavage and active sites of 434 repressor. We utilized the mutant repressors to show that the DNA-bound 434 repressor dimer overwhelmingly prefers to use an intramolecular mechanism of autocleavage. Our data suggest that the 434 repressor cannot be forced to use an intermolecular cleavage mechanism. Based on these data, we propose a model in which the cleavage-competent conformation of the repressor is stabilized by operator binding.
The life cycle of temperate lambdoid bacteriophage alternates between the lysogenic and lytic developmental pathways (9). In a lysogen, the phage's genome is integrated into the chromosome of its host and is replicated along with the host chromosome. The lysogenic pathway is a metastable developmental stage; all lysogenized phage can undergo lytic development (35). In lytic growth, phage DNA is not integrated into the chromosome; rather, its intracellular replication, assembly into phage particles, and subsequent host cell lysis result in phage production. The switch from lysogenic to lytic growth is governed by the activities of the phage's cI repressor protein. The repressor binds to specific operator regions on the phage DNA. Specific DNA binding allows the repressor to simultaneously activate transcription of genes essential for the maintenance of lysogeny and repress transcription of genes needed for lytic growth. Hence, inactivation of repressor DNA binding and its resulting regulatory activities leads to derepression of lytic genes and lytic growth (9, 10, 38).
Repressor inactivation and the resulting phage induction can occur when a host is treated with a DNA-damaging agent (15). This treatment leads to activation of the SOS response, the primary function of which is the repair of damaged DNA. The first step in activation of the SOS response is the formation of active RecA (RecA*), which stimulates autoproteolysis of LexA (18, 24, 25). Similarly, RecA* also stimulates autoproteolysis of the lambdoid phage repressors (10, 33, 38). In both cases, RecA*-stimulated autoproteolysis destroys the protein's abilities to bind DNA, leading to a loss of its gene-regulatory activities (22). Due to the destruction of the lambdoid phage repressor by the activity of RecA* during SOS, the lytic pathway, once entered, is irreversible.
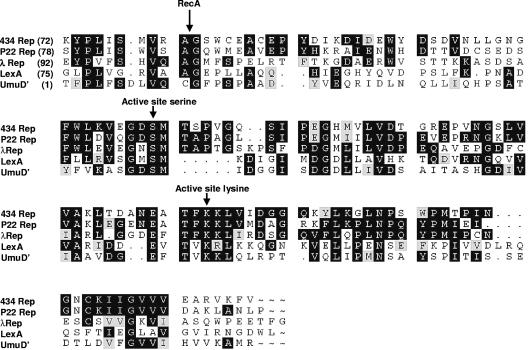
The organization of LexA and the organization of the lambdoid bacteriophage repressors are very similar. The N-terminal domains of these proteins make all of the specific and nonspecific contacts with DNA (40). Specific DNA binding by these proteins to an operator site requires dimerization of two protein monomers (19). In all of these proteins, the C-terminal domain is responsible for stable dimer formation, and in the phage repressors, the C-terminal domain mediates the formation of higher-order oligomers (4, 10, 31, 38). A third region, known as the “linker,” connects the two stably folded structural domains of the repressors (31, 39). The sequences and structures of the C-terminal domains of these proteins are similar to those of the bacterial UmuD protein (15) (Fig. 1). UmuD is a non-DNA-binding regulator of the SOS-dependent error-prone DNA synthesis activity (14, 44, 45). The activation of UmuD also requires RecA-stimulated autoproteolysis of the molecule (30).
FIG. 1.
Alignment of the sequences of the linkers and C-terminal domains of bacteriophage 434, P22, and λ repressors with the homologous regions of LexA and UmuD. The black background indicates positions where identical amino acids are found in at least three of the aligned sequences. The gray background indicates amino acids in the third and/or fourth position that are homologous to two identical residues. The alignment begins at the residues indicated. The positions of RecA cleavage and the active site serine and lysine are indicated by arrows.
How does RecA-stimulated autoproteolysis alter the activities of these proteins? In all these proteins, the “active site” residues responsible for catalyzing the autoproteolysis reaction are located in their C-terminal domains. The linker region contains a specific Ala (or Cys)-Gly cleavage site (however see reference 21) that is the target of C-terminal domain-catalyzed proteolysis (18, 23). Cleavage in the linker region separates the N-terminal portions of these proteins from the C-terminal domain. In the case of LexA and the lambdoid phage repressors, separation of the N-terminal domain from the C-terminal domain lowers the DNA affinity of the N-terminal domain, causing it to dissociate from its binding sites. This in turn eliminates the protein's ability to control transcription and, in the case of the bacteriophage repressors, leads to irreversible induction of the lytic pathway of the phage (3, 8, 10, 36, 38).
LexA, UmuD, and the phage repressors all function as dimers. However, LexA and the λ repressor cleave efficiently as monomers. Therefore, normally, the active site residues of one subunit of LexA and the λ repressor catalyze cleavage of the linker located within the same subunit; that is, in these proteins, cleavage must occur intramolecularly (16, 17, 22, 34, 36). UmuD undergoes cleavage as a dimer, and in the presence of RecA, it uses an intermolecular cleavage strategy (28, 29). In this case, one monomer of the UmuD dimer cleaves in the linker of its partner subunit (43). Similar to the λ repressor LexA and UmuD, RecA* stimulates autocleavage of the 434 repressor (32). As we show below, it is likely that the 434 repressor uses the same chemical mechanism of autocleavage as UmuD, the λ repressor, and LexA. In contrast to the λ repressor and LexA, however, the 434 repressor's intrinsic autoproteolysis activity is most active when the repressor is bound as a dimer to operator DNA (32). In this respect, the 434 repressor is more similar to the distantly related non-DNA-binding homolog UmuD than to the closer relative the λ repressor. However, we do not know whether the 434 repressor uses an inter- or intramolecular cleavage mechanism.
To identify the mechanism of 434 repressor autocleavage, we examined the autocleavage activities of wild-type 434 repressors and mutants bearing changes at residues in the proposed cleavage and active sites. Using these tools, we were unable to force the 434 repressor to undergo detectable intermolecular autocleavage. Our results demonstrates that despite cleaving most efficiently as a DNA-bound dimer, the 434 repressor overwhelmingly prefers to use an intramolecular mechanism.
MATERIALS AND METHODS
Site-directed mutagenesis of the 434 repressor gene.
A QuickChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) was used with pGEM 434 (32) as a template to create plasmids that direct expression of repressors bearing mutations in the cleavage (G89A) and active (S126A) sites. pGEM 434 contains the gene coding for the 434 repressor, the expression of which is under control of the inducible tac promoter (32). The following primer combinations were used: for G89A, 5′-ATGGTTAGAGCCGCCTCGTGGTGTGAA-3′ and 5′-TTCACACCACGAGGCGGCTCTAACCAT-3′; and for S126A, 5′-GTTGAAGGTGACGCCATGACCTCACCT-3′ and 5′-AGGTGAGGTCATGGCGTCACCTTCAAC-3′. The resultant plasmids were transformed into competent Escherichia coli X-90 (7) and plated on LB agar/ampicillin plates. Plasmid DNA was purified from cells that overexpressed a protein of the desired size, and the presence of the mutation was confirmed by DNA sequencing. The plasmids containing the correct mutations were designated 434S126A and 434G89A.
Strains bearing either plasmid pGEM434S126A or plasmid pGEM434G89A were used for purification of the respective repressor proteins. Purification of each repressor was performed by using the protocol described previously (1, 46). Similarly, wild-type 434 repressor was purified and characterized as described previously (1, 46).
Since some of our experiments required the S126A and G89A mutant repressors to be expressed in the same cell, repressor-producing plasmids with compatible replication origins were constructed. pACYC184 (New England Biolabs, Beverly, MA) was used due to its compatibility with the plasmids described above. The DNA encoding the 434G89A mutant repressor, including its promoter, was excised from pGEM434G89A by PvuII cleavage and inserted into the Bstz17I site of pACYC184. Restriction mapping confirmed the presence and orientation of the G89A-encoding DNA fragment.
Subcloning 434 OR1 into pGEM434 derivatives.
The 434 OR1 sequence was placed into the pGEM434 expression vectors by isolating the ∼300-bp DNA fragment containing this sequence from the pOR1 plasmid (2) by PvuII digestion and ligating this fragment into plasmids pGEM434, pGEM434S126A, and pGEM434G89A that had been linearized at their unique BsaAI sites. Restriction mapping of the resulting plasmids was performed to confirm insertion and to define the orientation of the desired fragment.
Determination of equilibrium binding constants.
The affinity of each mutant repressor for operator DNA was determined by both filter binding and gel mobility shift assays as described previously (5, 6, 27). Binding site DNA was obtained by annealing complementary 48-base oligomers (Integrated DNA Technologies, Coralville, IA) encoding 434 OR1 and radiolabeled as described previously (27). The results of both filter binding and gel shift experiments were visualized using a Storm 860 imager (Amersham, Piscataway, NJ), and dissociation constants (KD) were obtained from nonlinear fitting of these data to a hyperbolic equation using Prism (GraphPad Software, San Diego, Calif.). All protein concentrations were corrected for activity as previously described (11, 12, 27).
Alkaline-induced autoproteolysis.
Autoproteolysis at high pH was performed using a modification of the protocol described by Kim and Little (20). The repressor was incubated in buffer containing 50 mM CAPS (N-cyclohexyl-3-aminopropanesulfonic acid)-NaOH (pH 10.5) and 50 mM KCl at 37°C for 24 h. The amounts of each protein are indicated below. Following incubation, the samples were added to Tris/Tricine loading dye (0.1 M Tris, pH 6.8, 1% sodium dodecyl sulfate (SDS), 20 mM 2-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue) and boiled for 5 min. The reaction products were resolved by electrophoresis on 15% SDS-polyacrylamide Tris-Tricine gels (41). The repressor and its fragments were visualized by chemiluminescent detection of Western blots by a Storm Imager using an ECL-Plus kit (both obtained from Amersham, Piscataway, NJ) with horseradish peroxidase secondary antibody.
RecA-mediated in vitro autoproteolysis.
Autoproteolyis experiments were performed using conditions identical to those described previously (32). The standard buffer used in all assays contained 50 mM KCl, 15 mM Tris (pH 7.5), 2 mM MgCl2, 0.1 mM EDTA, and 2 mM dithiothreitol. Briefly, active RecA (RecA*) filaments were preformed at a concentration that was fivefold higher than the desired concentration by mixing 1.25 μM RecA (Epicenter, Madison, WI), 5 mM γ-S-ATP, and 0.5 μM oligo(dT)20 (expressed as a concentration of the oligonucleotide) in standard reaction buffer and incubating the preparation at room temperature for 10 min. Appropriate amounts of 5× RecA* filaments were added to tubes containing 200 ng (0.3 μM) or 2,000 ng (3.0 μM) of repressor in the absence or presence of a twofold excess of OR1 and incubated at 37°C for 4 h. Cleavage products were resolved on Tris/Tricine gels as described above. The products of the reaction were visualized by Western blotting as described above.
RecA-mediated autoproteolysis of the 434 repressor in vivo.
RecA*-mediated autoproteolysis in vivo was monitored using a method similar to the method described by McDonald et al. (28, 29). E. coli X-90 (7), transformed with the desired repressor-expressing plasmid(s), was grown to the mid-log phase (∼1.0 × 108 CFU/ml) in LB medium supplemented with the appropriate antibiotic(s). A portion of the culture was removed as an uninduced control. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the remaining culture at a final concentration of 2 μM to induce expression of the repressor protein(s). After 1 h of induction another sample was removed. Mitomycin C was added to the remaining culture at a final concentration of 3.5 μg/ml to initiate the SOS response. Sixty minutes after mitomycin C addition, a sample was taken. Control experiments established that the maximal amount of repressor cleavage was obtained within this time. Each sample was spun at 10,000 × g for 3 min to pellet the cells. The supernatants were removed, and the pelleted cells were lysed in 1 ml of Tris/Tricine loading buffer. The crude extracts were fractionated by electrophoresis through 15% Tris/Tricine SDS-polyacrylamide gels, the cleavage products were transferred to polyvinylidene difluoride membranes, and the products were visualized by Western blotting as described above.
RESULTS
To determine whether the 434 repressor utilizes an inter- or intramolecular autocleavage mechanism, we used an approach similar to the approach used by Kim and Little (20) and McDonald et al. (28, 29) and created two repressor mutants having substitutions in the residues that comprise part of either the active (or “catalytic”) site of the 434 repressor (S126) or its cleavage site (G89). We examined the ability of each protein to undergo autocleavage alone, in combination with the other protein, or in the presence of the wild-type repressor. Since the cleavage and active site residues were identified by homology to the residues found in the related λ and LexA repressors and other proteins (Fig. 1), we first verified that these residues play a role in repressor autocleavage. Regardless of whether the repressor uses an inter- or intramolecular autocleavage mechanism, the S126A and G89A mutant proteins should be incapable of undergoing autocleavage under any conditions. This is because the S126A repressor mutation eliminates a residue hypothesized to catalyze peptide bond cleavage and the G89A change is hypothesized to destroy the “substrate” (cleavage) site. We tested this hypothesis by comparing the abilities of the wild-type repressor and the two mutant repressors to self-cleave. We detected autoproteolysis by probing Western blots of the gel-separated reaction products with antirepressor antibodies. We chose to detect the cleavage products by Western blotting as this method is >100 times more sensitive than Coomassie blue staining (see Discussion).
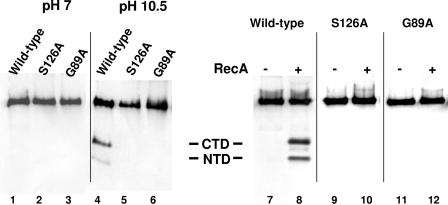
Compared to reactions in which the wild-type repressor was incubated at neutral pH (Fig. 2, lane 1), incubating 200 ng (0.3 μM) 434 repressor at pH 10.5 resulted in the appearance of two lower-molecular-weight antibody-reactive species (Fig. 2, lane 4). The molecular weights of these products correspond to the molecular weights of the repressor N- and C-terminal domain fragments. This finding shows that similar to the λ repressor and LexA (20, 22), the wild-type 434 repressor self-cleaves at a high pH in vitro. In contrast, no lower-molecular-weight antibody reactive products were observed when 200 ng of either the G89A or S126A mutant repressor was incubated under the same conditions (Fig. 2, lanes 2, 3, 5, and 6). We also examined the ability of activated RecA to stimulate autocleavage of the wild-type, S126A, and G89A repressors at physiological pH (Fig. 2, lanes 7 to 12). Similar to the results obtained at high pH, only the wild-type repressor (Fig. 2, lanes 7 and 8) and not the S126A (Fig. 2, lanes 9 and 10) or G89A (Fig. 2, lanes 11 and 12) mutant repressor underwent autocleavage in the presence of active RecA. These findings are consistent with our hypothesis that S126 and G89 each play a role in mediating repressor autocleavage.
FIG. 2.
In vitro autoproteolysis of the wild-type 434 repressor and the active (434S126A) and cleavage (434G89A) site mutant derivatives. The wild-type 434 repressor (200 ng) and its mutant derivatives were incubated in the absence of RecA at pH 7 (lanes 1 to 3) or pH 10.5 (lanes 4 to 5) for 24 h or in the absence (lanes 7, 9, and 11) or presence (lanes 8,10, 12) of active RecA at pH 7.5 (see Materials and Methods) for 4 h. Intact repressors and the cleavage products were fractionated on 15% Tris/Tricine gels and visualized by Western blotting using anti-434 repressor antibodies. The positions of the N- and C-terminal cleavage products are indicated in the middle. CTD, C-terminal domain; NTD, N-terminal domain.
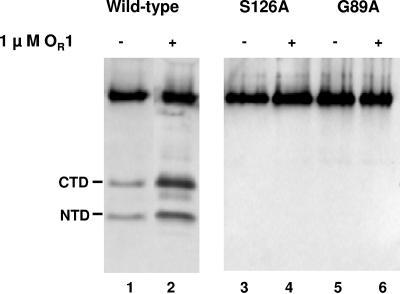
Rather than the monomeric form, the dimeric, operator-bound conformation of the 434 repressor is the form of repressor that most efficiently undergoes autoproteolysis in the presence of RecA (32). Thus, we hypothesized that the presence of DNA may stimulate autocleavage of the S126A and G89A mutant repressors. Hence, we examined the ability of active RecA to stimulate autocleavage of the wild-type, S126A, and G89A repressors at physiological pH in the presence of excess operator DNA (Fig. 3). Similar to the results obtained previously, added DNA markedly stimulated autocleavage of the wild-type repressor (Fig. 3, lanes 1 and 2). However, neither the S126A mutant repressor (Fig. 3, lanes 3 and 4) nor the G89A mutant repressor (Fig. 3, lanes 5 and 6) underwent autocleavage in the presence of active RecA and operator DNA.
FIG. 3.
Effect of added DNA on the ability of the wild-type 434 repressor and the active (434S126A) and cleavage (434G89A) site mutant derivatives of the 434 repressor to undergo intermolecular autocleavage. The wild-type 434 repressor (200 ng) and its mutant derivatives were incubated in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of excess 434 OR1 DNA. Intact repressors and the cleavage products were fractionated on 15% Tris/Tricine gels and visualized by Western blotting using anti-434 repressor antibodies. The positions of the N- and C-terminal cleavage products are indicated on the left. CTD, C-terminal domain; NTD, N-terminal domain.
It is possible that the failure of the S126A and G89A mutant repressors to undergo efficient autocleavage resulted from a mutation-induced change in repressor conformation that altered the overall structure and thus the autocleavage ability. To begin to ascertain whether the S126A or G89A mutation affects the overall repressor structure, the affinities of these proteins for the naturally occurring 434 repressor-binding site, OR1, were determined. Compared to the wild-type 434 repressor, the S126A and G89A repressor mutations decreased the affinity of the proteins for OR1 by only four- and twofold, respectively (Table 1). This finding shows that the S126A and G89A mutations affect the operator binding of the 434 repressor only slightly. Since dimerization of the 434 repressor is a prerequisite for its DNA binding, the small effect of a mutation on the affinity of the repressor for OR1 indicates that the S126A and G89A mutations do not dramatically alter repressor dimer formation. Moreover, cells that synthesize either the wild-type or mutant repressors at a low level were immune to infection by a cI− 434 bacteriophage, indicating that the mutant proteins are functional in vivo (data not shown). Taken together, Table 1 and Fig. 2 and 3 indicate that the S126A and G89A mutations inhibit repressor autocleavage by altering residues that participate directly in the cleavage reaction, not by altering repressor structure or function.
TABLE 1.
Affinities of the wild-type 434 repressor and two noncleavable mutant repressors for 434 OR1
| Repressor variant | KD(M)a |
|---|---|
| Wild-type 434 repressor | 2 × 10−9 (0.8 × 10−9) |
| S126A | 8 × 10−9 (0.9 × 10−9) |
| G89A | 5 × 10−9 (0.4 × 10−9) |
Equilibrium dissociation constants were calculated from binding assays as described in Materials and Methods.The values in parentheses are standard deviations.
UmuD and MucA proteins having mutations at residues homologous to S126 and G89 are incapable of undergoing RecA*-stimulated autocleavage (29). However, cleavage of UmuD and MucA can take place when a subunit having a mutation in the active site is mixed with a subunit having a mutation in the cleavage site (29). This observation demonstrates that UmuD and MucA are capable of using an intermolecular mechanism of autocleavage. Using the S126A and G89A repressors, we pursued a similar approach to probe the mechanism of 434 repressor autocleavage.
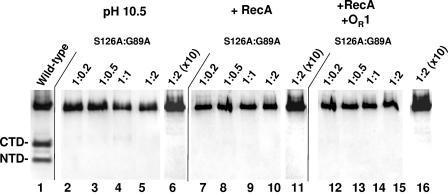
We examined the ability of the G89A repressor, the repressor variant having a functional active site, to cleave the S126A repressor, which has an intact cleavage site, at high pH. To increase the sensitivity of the reaction, we varied the ratio of the enzyme (G89A) to the substrate (S126A). Regardless of the ratio of these proteins, we could not detect any products indicative of repressor autocleavage (Fig. 4, lanes 2 to 5). To determine whether the failure to identify autoproteolysis was due to the inability of the Western blotting to detect weak autocleavage, we repeated the experiments whose results are shown in Fig. 4, lanes 2 to 5, with a 10-fold-higher total concentration of repressor. A sample of the results obtained at a 2:1 ratio of S126A to G89 is shown in Fig. 4, lane 7. Under these conditions, we were unable to detect any repressor autocleavage products. These findings suggest that the 434 repressor can undergo autoproteolysis only by an intramolecular cleavage mechanism.
FIG. 4.
pH- and RecA-stimulated cleavage of mixtures of cleavage site (G89A) and active site (S126A) mutant 434 repressors. The wild-type repressor (200 ng) was incubated with active RecA for 4 h (lane 1) as a control. The cleavage site (G89A) and active site (S126A) mutant 434 repressors were mixed in various ratios (lanes 2 to 5, 7 to 10, and 12 to 15 contained 200 ng of 434S126A plus [434G89A; lanes 6, 11, and 16 contained 2,000 ng of 434S126A plus 434G89A) and were incubated at pH 10.5 for 24 h (lanes 2 to 6), in the presence of RecA for 4 h (lanes 7 to 11), or in the presence of RecA and a twofold molar excess of 434 OR1 for 4 h (lanes 12 to 16). Intact repressors and the cleavage products were fractionated on 15% Tris/Tricine gels and visualized by Western blotting using anti-434 repressor antibodies. The positions of the intact repressor and the N- and C-terminal cleavage products are indicated on the left. CTD, C-terminal domain; NTD, N-terminal domain.
We also examined whether the G89A mutant repressor was able to cleave the S126A protein when active RecA was used to stimulate the cleavage reaction. We found that under conditions in which efficient RecA-stimulated autocleavage of the wild-type repressor occurred (Fig. 2) incubating the G89A repressor with the S126A repressor did not result in the formation of any repressor autocleavage products at lower (Fig. 4, lanes 7 to 10) or higher (Fig. 4, lane 11) repressor concentrations. Similarly, we found that adding excess OR1 DNA, which stimulated cleavage of the wild-type repressor (Fig. 3), did not facilitate cleavage of the mixtures of G89A and S126A mutant repressors at any concentration (Fig. 4, lanes 12 to 16).
Repressor autocleavage in vivo.
All of the results presented above suggesting that the 434 repressor uses only an intramolecular cleavage strategy were performed in vitro using purified proteins. The studies demonstrating that UmuD can utilize an intermolecular autocleavage mechanism were based on analysis of in vivo-generated cleavage products (28). Hence, it is possible that our in vitro reactions lacked a factor(s) that could allow the 434 repressor to utilize an intermolecular autocleavage mechanism. To test this idea, we compared the results of two autocleavage experiments. In one experiment, we monitored autocleavage of the wild-type repressor under a variety of conditions in a bacterial strain that expresses only this protein. In the second experiment, we monitored repressor autocleavage in an E. coli strain that contains two plasmids, one directing the expression of the G89A (cleavage site) mutant 434 repressor and the other directing the expression of the S126A (active site) mutant repressor. RecA*-mediated autocleavage of the repressor variants was stimulated by addition of mitomycin C to the strains. We visualized the repressors and their cleavage products by Western blotting using antibodies directed against the intact 434 repressor (see above).
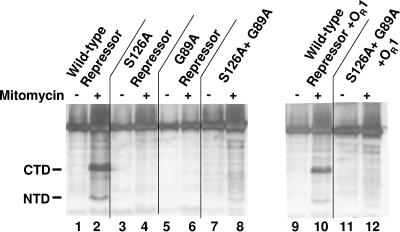
Upon addition of 2 μM IPTG to cells bearing the plasmid that directs expression of the 434 repressor, our antibodies detected the presence of the 434 repressor (Fig. 5, lane 1). When these cells were incubated with IPTG and mitomycin C, two additional lower-molecular-weight antirepressor antibody-reactive species were also detected (Fig. 5 lane 2). The molecular weights of these species correspond to those of the N- and C-terminal domains of the 434 repressor. Since the C-terminal domain is stable in vivo (4, 11), we used the data in Fig. 5 to estimate the amount of repressor cleavage that occurred in vivo under these conditions. A comparison of the amounts of the C-terminal domain and the intact repressor indicated that ∼35% of the 434 repressor was cleaved into the component domains under these conditions. This finding shows that mitomycin C-dependent SOS induction stimulates autocleavage of the wild-type 434 repressor in vivo.
FIG. 5.
Autocleavage of the 434 repressor in vivo. Mid-log-phase cells containing plasmids that direct the expression of the wild-type and/or mutant repressors were induced for 1 h with 2 μM IPTG and grown for an additional 1 h in the absence or presence of 3.5 μg of mitomycin C and then processed for electrophoresis. The blot is a Western blot of the resulting products probed with antibodies directed against the 434 repressor. Cells contained only nonspecific DNA (lanes 1 to 8) or 434 OR1 (lanes 9 to 12). The positions of the SOS-induced repressor fragments are indicated on the left (NTD, amino terminal domain; CTD, carboxyl-terminal domain).
In contrast to the results obtained with cells expressing the wild-type 434 repressor, addition of mitomycin C to strains that expressed either the S126A repressor (Fig. 5, lanes 3 and 4) or the G89A repressor (Fig. 5, lanes 5 and 6) or both of these proteins (Fig. 5, lanes 7 and 8) did not result in the production of any SOS-dependent repressor cleavage products. This finding is consistent with the in vitro data indicating that the 434 repressor uses an intramolecular autocleavage mechanism.
Since 434 autocleaves most efficiently as the DNA-bound dimer, we tested whether the presence of specific DNA binding sites in vivo affected the autocleavage wild-type repressor and the mixtures of G89A and S26A repressors. To do this, we placed 434 OR1 into the plasmid that overexpressed the wild-type repressor or the S126A repressor and examined the effect of mitomycin C addition on repressor autocleavage. Addition of mitomycin C to cells bearing the wild-type repressor and 434 OR1 effectively induced wild-type repressor autocleavage (Fig. 5, lanes 9 and 10). Similar to the results obtained in vitro, the presence of 434 OR1 was not sufficient to stimulate SOS-dependent repressor cleavage in cells that produced both the G89A repressor and the S126A repressor (Fig. 5, lanes 11 and 12). Thus, under no circumstances (either in vitro or in vivo) were we able to detect intermolecular cleavage of the 434 repressor.
DISCUSSION
The data presented above indicate that the 434 repressor overwhelmingly prefers to undergo autoproteolysis using an intramolecular mechanism. The other DNA binding repressors, such as LexA and the λ repressor, also preferentially utilize intramolecular cleavage (42). However, 434 repressor's autocleavage mechanism differs from the mechanisms of these other repressors. The other repressors undergo efficient autocleavage as monomers. In contrast, the 434 repressor undergoes autocleavage most efficiently as a DNA-bound dimer (32). With respect to the oligomerization state of the autocleavage substrate, the 434 repressor resembles UmuD. Dimerization of UmuD facilitates the use of an intermolecular mechanism of autocleavage (28), but in the case of the 434 repressor, dimerization facilitates intramolecular autocleavage. Hence, although LexA, the λ repressor, UmuD, and the 434 repressor are homologous proteins and appear to use the same chemical cleavage mechanism, the quaternary structural requirements and the reaction order needed for implementation of this mechanism are different for all these proteins.
One question that stems from our findings is whether the 434 repressor can use an intermolecular autocleavage mechanism under any circumstances. The answer to this question depends on whether our methodology is sensitive enough to detect the apparently highly unfavorable intermolecular cleavage event in mixtures of the G89A and S126A mutant repressors (Fig. 3 to 5). In our standard reaction conditions, 40 to 50% of the input wild-type repressor undergoes autocleavage either in the presence of RecA or upon incubation at alkaline pH (Fig. 2). In either case, an extended incubation time does not increase the amount of repressor cleaved (data not shown), nor did we see >50% cleavage of repressor in vivo. These findings are consistent with data showing that the tight complex that forms between the intact repressor and its C-terminal domain is resistant to autocleavage (Koudelka, unpublished results). Thus, our conditions adequately sample the maximal extent of repressor cleavage. Dilution experiments revealed that our Western blotting procedure can detect cleavage products at amounts that are 1/300 (∼0.6 ng) of the amount formed in our standard in vitro cleavage experiments. This observation, combined with the finding that we detected no cleavage products in reactions with 10-fold more repressor than our usual experiment (2,000 ng of total repressor) (Fig. 4, lanes 9, 14, and 19), suggests that if intermolecular cleavage of the 434 repressor occurs, the rate for this type of cleavage mechanism must be ≥1,000-fold lower than the rate of intramolecular cleavage. Hence, we concluded that while it may be possible for the 434 repressor to utilize an intermolecular cleavage strategy, the rate of this cleavage is so low that the 434 repressor should be considered incapable of undergoing autocleavage using an intermolecular mechanism.
Having established that each monomer in the DNA-bound repressor dimer cleaves itself, we are interested in understanding why this form of repressor is the form that most efficiently undergoes autocleavage. To answer this question, we considered several observations concerning the effect of DNA on repressor structure and function. First, DNA binding-induced repressor dimerization allosterically alters the conformation of the repressor in the vicinity of the linker polypeptide (12). Together with fluorescence data on our tryptophan mutant repressors (12), model-building studies indicated that this conformation change occurs in the region between amino acids ∼80 to 110, a portion of the repressor polypeptide that surrounds the cleavage site. The nature of fluorescence changes indicates that this region becomes more tightly associated with the body of the C-terminal domain when the repressor binds DNA (12). Second, concentration-induced repressor dimerization only weakly affects the conformation of this region of the protein (12) and only weakly stimulates repressor autocleavage. Based on these findings, we suggest that the juxtaposition of the cleavage site with the active site in a DNA-bound dimer depends on a DNA binding-driven conformational change in the region of the repressor that surrounds the cleavage site and that the association of this region with the body of the repressor is required for autocleavage. We speculate that under any circumstances the “affinity” of the linker “substrate” for the “active” site pocket on an opposite subunit is too low to permit efficient cleavage. This lowered affinity may be due to steric constraints imposed by the sequence of the linker region, or the strength of the interactions may be too weak to support intermolecular interactions. In either case, the weak interaction would prohibit the repressor from utilizing an intermolecular cleavage mechanism. The idea that the conformation of the polypeptide surrounding the linker can affect cleavage is consistent with the finding that mutations in this region in LexA and the λ repressor can increase the rate of autocleavage of these proteins (16, 20).
Our proposed explanation for why the 434 repressor undergoes autocleavage most efficiently as a DNA-bound dimer includes features of the models that describe the autocleavage strategies of two other self-cleaving proteins, LexA and UmuD. Thus, our proposed mechanism for 434 repressor autoproteolysis is an elaboration of the mechanism proposed for these proteins. Analogous to the conformational change observed upon DNA binding by the 434 repressor, structural studies indicate that in UmuD, in order to undergo autocleavage, the first ∼30 residues of this protein, which includes the cleavage site, must undergo a large a conformational change when the protein associates with RecA* (13). This conformational change allows this region of each monomer to loop back into the catalytic domain of its dimer partner (13), thereby facilitating intermolecular cleavage.
The current model of LexA autoproteolysis proposes that in the absence of RecA, LexA equilibrates between two conformations, a cleavage-competent form and an incompetent form (26). The model proposes that the cleavage-incompetent conformation predominates under normal physiological conditions. However when the molecule is associated with RecA, the cleavage-competent conformation is stabilized and becomes the predominant conformation (37). In this form, the linker is juxtaposed within a short distance from the catalytic site, creating a conformation that is readily cleaved. Our findings with the 434 repressor fit into a similar conformation-equilibrium model in which operator binding places the repressor molecules in the protein-DNA complex into a cleavage-incompetent conformation that is more likely to undergo autoproteolysis when it is presented to RecA.
Acknowledgments
This work was supported by grant MCB-0239000 from the National Science Foundation.
We thank members of our laboratory and Mark Sutton for critical comments on the work and manuscript.
REFERENCES
- 1.Anderson, J. E., M. Ptashne, and S. C. Harrison. 1984. Co-crystals of the DNA-binding domain of phage 434 repressor and a synthetic 434 operator. Proc. Natl. Acad. Sci. USA 81:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, A. C., and G. B. Koudelka. 1993. Operator sequence context influences amino acid-base-pair interactions in 434 repressor-operator complexes. J. Mol. Biol. 234:542-553. [DOI] [PubMed] [Google Scholar]
- 3.Bushman, F. D. 1993. The bacteriophage 434 right operator. Roles of OR1, OR2 and OR3. J. Mol. Biol. 230:28-40. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, P. A., and G. B. Koudelka. 1994. Expression, purification, and functional characterization of the carboxyl-terminal domain fragment of bacteriophage 434 repressor. J. Bacteriol. 176:6907-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciubotaru, M., F. V. Bright, C. M. Ingersoll, and G. B. Koudelka. 1999. DNA-induced conformational changes in bacteriophage 434 repressor. J. Mol. Biol. 294:859-873. [DOI] [PubMed] [Google Scholar]
- 6.Ciubotaru, M., and G. B. Koudelka. 2003. DNA stimulated assembly of oligomeric bacteriophage 434 repressor: evidence for cooperative binding by recruitment. Biochemistry 42:4253-4264. [DOI] [PubMed] [Google Scholar]
- 7.Coulandre, C., and J. H. Miller. 1977. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol. 117:525-567. [DOI] [PubMed] [Google Scholar]
- 8.Craig, N. L., and J. W. Roberts. 1980. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature 283:26-30. [DOI] [PubMed] [Google Scholar]
- 9.Daniels, D. L., J. L. Schroeder, W. Szybalski, F. Sanger, A. R. Coulson, G. F. Hong, D. F. Hill, G. F. Petersen, and F. R. Blattner. 1983. Lambda II, p. 519-676. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 10.DeAnda, J., A. R. Poteete, and R. T. Sauer. 1983. P22 c2 repressor-domain structure and function. J. Biol. Chem. 258:10536-10542. [PubMed] [Google Scholar]
- 11.Donner, A. L., P. A. Carlson, and G. B. Koudelka. 1997. Dimerization specificity of P22 and 434 repressors is determined by multiple polypeptide segments. J. Bacteriol. 179:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donner, A. L., and G. B. Koudelka. 1998. Carboxyl-teminal domain dimer interface mutant 434 repressors have altered dimerization and DNA binding specificities. J. Mol. Biol. 283:931-946. [DOI] [PubMed] [Google Scholar]
- 13.Ferentz, A. E., T. Opperman, G. C. Walker, and G. Wagner. 1997. Dimerization of the UmuD′ protein in solution and its implications for regulation of SOS mutagenesis. Nat. Struct. Biol. 4:979-983. [DOI] [PubMed] [Google Scholar]
- 14.Ferentz, A. E., G. C. Walker, and G. Wagner. 2001. Converting a DNA damage checkpoint effector (UmuD2C) into a lesion bypass polymerase (UmuD′2C). EMBO J. 20:4287-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 16.Gimble, F. S., and R. T. Sauer. 1986. Lambda repressor inactivation: properties of purified ind- proteins in the autodigestion and RecA-mediated cleavage reactions. J. Mol. Biol. 192:39-47. [DOI] [PubMed] [Google Scholar]
- 17.Gimble, F. S., and R. T. Sauer. 1989. Lambda repressor mutants that are better substrates for RecA-mediated cleavage. J. Mol. Biol. 206:29-39. [DOI] [PubMed] [Google Scholar]
- 18.Horii, T., T. Ogawa, T. Nakatani, T. Hase, H. Matsubara, and H. Ogawa. 1981. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell 27:515-522. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, A. D., C. O. Pabo, and R. T. Sauer. 1982. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 65:839-856. [DOI] [PubMed] [Google Scholar]
- 20.Kim, B., and J. W. Little. 1993. LexA and lambda Cl repressors as enzymes: specific cleavage in an intermolecular reaction. Cell 73:1165-1173. [DOI] [PubMed] [Google Scholar]
- 21.Koudelka, A. P., L. A. Hufnagel, and G. B. Koudelka. 2004. Purification and characterization of the repressor of the Shiga toxin-encoding bacteriophage 933W: DNA binding, gene regulation, and autocleavage. J. Bacteriol. 186:7659-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. USA 81:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 24.Little, J. W., S. H. Edmiston, L. Z. Pacelli, and D. W. Mount. 1980. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc. Natl. Acad. Sci. USA 77:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little, J. W., D. W. Mount, and C. R. Yanisch-Perron. 1981. Purified lexA protein is a repressor of the recA and lexA genes. Proc. Natl. Acad. Sci. USA 78:4199-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, Y., R. A. Pfuetzner, S. Mosimann, M. Paetzel, E. A. Frey, M. Cherney, B. Kim, J. W. Little, and N. C. Strynadka. 2001. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell 106:585-594. [DOI] [PubMed] [Google Scholar]
- 27.Mauro, S. A., D. Pawlowski, and G. B. Koudelka. 2004. The role of the minor groove substituents in indirect readout of DNA sequence by 434 repressor. J. Biol. Chem. 278:12955-12960. [DOI] [PubMed] [Google Scholar]
- 28.McDonald, J. P., E. G. Frank, A. S. Levine, and R. Woodgate. 1998. Intermolecular cleavage by UmuD-like mutagenesis proteins. Proc. Natl. Acad. Sci. USA 95:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald, J. P., T. S. Peat, A. S. Levine, and R. Woodgate. 1999. Intermolecular cleavage by UmuD-like enzymes: identification of residues required for cleavage and substrate specificity. J. Mol. Biol. 285:2199-2209. [DOI] [PubMed] [Google Scholar]
- 30.Nohmi, T., J. R. Battista, L. A. Dodson, and G. C. Walker. 1988. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc. Natl. Acad. Sci. USA 85:1816-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pabo, C. O., R. T. Sauer, J. M. Sturtevant, and M. Ptashne. 1979. The lambda repressor contains two domains. Proc. Natl. Acad. Sci. USA 76:1608-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawlowski, D. R., and G. B. Koudelka. 2004. The preferred substrate for RecA-mediated cleavage of bacteriophage 434 repressor is the DNA-bound dimer. J. Bacteriol. 186:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phizicky, E. M., and J. W. Roberts. 1980. Kinetics of RecA protein-directed inactivation of repressors of phage lambda and phage P22. J. Mol. Biol. 139:319-328. [DOI] [PubMed] [Google Scholar]
- 34.Phizicky, E. M., and J. W. Roberts. 1981. Induction of SOS functions: regulation of proteolytic activity of E. coli RecA protein by interaction with DNA and nucleoside triphosphate. Cell 25:259-267. [DOI] [PubMed] [Google Scholar]
- 35.Ptashne, M. 1986. A genetic switch. Blackwell Press, Palo Alto, CA.
- 36.Roberts, J. W., C. W. Roberts, N. L. Craig, and E. M. Phizicky. 1979. Activity of the Escherichia coli recA-gene product. Cold Spring Harbor Symp. Quant. Biol. 43:917-920. [DOI] [PubMed] [Google Scholar]
- 37.Roland, K. L., M. H. Smith, J. A. Rupley, and J. W. Little. 1992. In vitro analysis of mutant LexA proteins with an increased rate of specific cleavage. J. Mol. Biol. 228:395-408. [DOI] [PubMed] [Google Scholar]
- 38.Sauer, R. T., H. C. Nelson, K. Hehir, M. H. Hecht, F. S. Gimble, J. DeAnda, and A. R. Poteete. 1983. The lambda and P22 phage repressors. J. Biomol. Struct. Dyn. 1:1011-1022. [DOI] [PubMed] [Google Scholar]
- 39.Sauer, R. T., M. J. Ross, and M. Ptashne. 1982. Cleavage of the lambda and P22 repressors by recA protein. J. Biol. Chem. 257:4458-4462. [PubMed] [Google Scholar]
- 40.Sauer, R. T., R. R. Yocum, R. F. Doolittle, M. Lewis, and C. O. Pabo. 1982. Homology among DNA-binding proteins suggests use of a conserved supersecondary structure. Nature 298:447-451. [DOI] [PubMed] [Google Scholar]
- 41.Schagger, H., and G. Von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 42.Slilaty, S. N., J. A. Rupley, and J. W. Little. 1986. Intramolecular cleavage of LexA and phage lambda repressors: dependence of kinetics on repressor concentration, pH, temperature, and solvent. Biochemistry 25:6866-6875. [DOI] [PubMed] [Google Scholar]
- 43.Sutton, M. D., A. Guzzo, I. Narumi, M. Costanzo, M. Altenbach, A. E. Ferentz, W. Hubbell, and G. C. Walker. 2002. A model for the structure of the Escherichia coli SOS-regulated UmuD2 protein. DNA Repair 1:77-93. [DOI] [PubMed] [Google Scholar]
- 44.Tang, M., I. Bruck, R. Eritja, J. Turner, E. G. Frank, R. Woodgate, M. O'Donnell, and M. F. Goodman. 1998. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc. Natl. Acad. Sci. USA 95:9755-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wharton, R. P., E. L. Brown, and M. Ptashne. 1985. Substituting an α-helix switches the sequence specific DNA interactions of a repressor. Cell 38:361-369. [DOI] [PubMed] [Google Scholar]