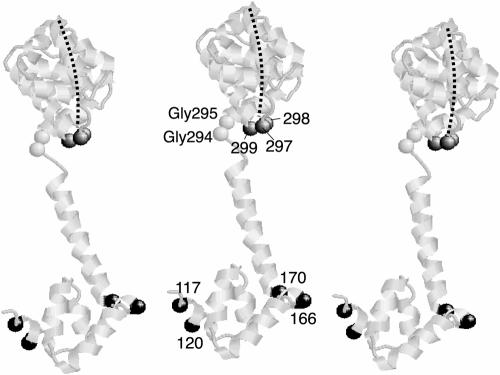
FIG. 8.
Model for the arrangement of FliG subunits. Three copies of the FliGMC protein from T. maritima are shown. The Gly-Gly motif and the positions giving high-yield cross-linking are indicated by space-filled alpha carbons and E. coli residue numbers. The FliGM domain is oriented with residues 117 and 120 of one subunit near residues 166 and 170 of the adjacent subunit. The FliGC domain is positioned with the active site ridge (indicated by the dashed line) toward the viewer and oriented in an approximately radial direction relative to the axis of the flagellum. The relative orientation of FliGM and FliGC domains is somewhat different from that in the crystal structure and was adjusted by making sterically allowed changes in torsion angles of the Gly-Gly linker. The three positions in FliGC that gave a cross-linked dimer with a moderate yield are also indicated. One of the residues in FliGC that gave a cross-linked dimer (residue 297 in E. coli; residue 299 in T. maritima) is at the end of the charge-bearing ridge and is among the charged residues important for function.