Abstract
With few novel antimicrobials in development, resistance to the current selection of antibiotics increasingly encroaches on our ability to control microbial infections. One limitation in our understanding of the basis of the constraints on current therapies is our poor understanding of antibiotic interactions with bacteria on a global scale. Custom DNA microarrays were used to characterize the response of Pseudomonas aeruginosa to ciprofloxacin, a fluoroquinolone commonly used in therapy against chronic infections by this intrinsically resistant bacterium. Of the approximately 5,300 open reading frames (ORFs) on the array, 941 genes showed statistically significant (P ≤ 0.05) differential expression in response to 0.3× MIC of ciprofloxacin; 554 were promoted and 387 were repressed. Most striking among the responsive genes was the region between PA0613 and PA0648, which codes for the bacteriophage-like R2/F2 pyocins. In this region, virtually every ORF was increased by 0.3× MIC of ciprofloxacin and even more dramatically up-regulated (7- to 19-fold) following treatment with 1× MIC of ciprofloxacin. Pyocin gene expression was confirmed with lux reporter mutants and real-time PCR studies; pyocin-like particles were also present in transmission electron micrographs of supernatants from cells treated with 1× MIC of ciprofloxacin. Interestingly, mutants in this region exhibited ≥8-fold-increased resistance to ciprofloxacin and other fluoroquinolones, demonstrating that this region is a susceptibility determinant. Since this region is known to be variably present in the genomes of clinical isolates of P. aeruginosa (R. K. Ernst et al., Environ. Microbiol. 5:1341-1349, 2003, and M. C. Wolfgang et al., Proc. Natl. Acad. Sci. USA 100:8484-8489, 2003), these findings demonstrate that the R2/F2 pyocin region is a “loaded gun” that can mediate fluoroquinolone susceptibility in P. aeruginosa.
Of particular concern in clinical settings is the development of “superbugs” which have acquired resistance to the vast majority of available antimicrobials, severely limiting treatment options and thus clinical outcomes. Pseudomonas aeruginosa is an important opportunistic pathogen because of its high intrinsic resistance to most antimicrobials used in therapeutic practice. This high intrinsic resistance is primarily due to a low outer membrane permeability coupled with various secondary resistance mechanisms (9), which effectively take advantage of the low antibiotic exposure afforded by the low permeability. Antibiotic exposure is further slowed by the propensity of the organism to become mucoid (23) and adopt a biofilm growth mode (2, 3), thereby permitting rapid mutation of the organism to a clinically relevant resistance state (8) and earning Pseudomonas “superbug” status.
In the absence of novel antimicrobials, an ability to control the development of antibiotic resistance to the current repertoire of antibiotics is critical. However, we are limited by our lack of knowledge of the interactions with pathogens and mechanisms of action of antimicrobial agents. This lack of knowledge constrains current therapies and consequently the development of new antibiotics. In an attempt to better understand these factors, we utilized DNA microarray technology to investigate the influence of subinhibitory and inhibitory concentrations of ciprofloxacin on the transcriptome of P. aeruginosa. Similar microarray studies on the interactions of antibiotics with other medically important bacteria have defined expression patterns or “antimicrobial signatures” for these agents that in part reflect the mechanism of action of the antimicrobial being examined, as well as the pathogen being studied (7, 14, 22). Ciprofloxacin interaction with Haemophilus influenzae at inhibitory and suprainhibitory conditions, for example, was reported to mainly induce the expression of SOS and DNA repair systems, an expression pattern reflective of the organism's attempt to protect itself from the DNA damage effects of DNA gyrase inhibition (7). Analysis of antimicrobial signatures, however, can become complicated by the inhibition of secondary targets, as well as downstream effects that result from inhibition of the primary target (26); subinhibitory concentrations of ciprofloxacin were therefore used to reduce such complicating effects.
Ciprofloxacin remains a clinically relevant antimicrobial in the treatment of P. aeruginosa infections, particularly chronic cystic fibrosis infections (6), and is thought to be well characterized with respect to mechanism of action and resistance (12). In this study, we utilized a functional genomic approach to demonstrate that P. aeruginosa strain PAO1 has a previously unknown fluoroquinolone susceptibility determinant.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are described in Table 1. P. aeruginosa PAO1 strain H103 was used for most studies. In all experiments, overnight aerobic cultures were grown with agitation in Luria-Bertani (LB) broth (1.0% tryptone, 0.5% yeast extract, 5% NaCl) (Difco Laboratories, Detroit, MI) at 37°C and used to inoculate 50-ml portions of LB broth in 500-ml Erlenmeyer flasks. Cultures were either untreated or treated with 0.3× or 1× MIC of ciprofloxacin and grown to the mid-logarithmic phase (optical density at 600 nm, 0.5 to 0.6). All microarray experiments were performed on three independent cultures and repeated at least twice per culture sample. Ciprofloxacin hydrochloride was obtained from Bayer (United Kingdom) and made fresh daily in distilled water.
TABLE 1.
Pseudomonas aeruginosa strains used in this study
| Strain | Genotype | Abbreviation | Reference or source |
|---|---|---|---|
| H103 | Wild-type P. aeruginosa PAO1 | H103 | |
| 57 | PA0611::ISlacZ/hah derivative of PAO1 | prtR::lacZ | 15 |
| 6322 | PA0613::ISlacZ/hah derivative of PAO1 | PA0613::lacZ | 15 |
| PAO1_lux_22_E4 | PA0620::luxCDABE derivative of H103; Tcr | PA0620::lux | This study |
| 3501 | PA0621::ISlacZ/hah derivative of PAO1 | PA0621::lacZ | 15 |
| PAO1_lux_26_H2 | PA0641::luxCDABE derivative of H103; Tcr | PA0641::lux | This study |
| 43080 | PA3617::ISphoA/hah derivative of PAO1 | recA::phoA | 15 |
| PAO1_lux_24_A3 | PA3866::luxCDABE derivative of H103; Tcr | PA3866::lux | This study |
Determination of MICs.
The MICs of antibiotics for the various P. aeruginosa strains were measured using the broth microdilution technique (1). Antimicrobials were made fresh daily and were obtained from the following sources: enofloxacin (Warner Lambert Co., Ann Arbor, MI), gentamicin (ICN Biomedicals Inc., Aurora, Ohio), norfloxacin (Sigma), nalidixic acid (Sigma), cefepime (Bristol Myers Squibb), and amikacin (Bristol Laboratories, Belleville, Ontario, Canada).
Microarray processing and data analysis.
RNA was isolated using QMIDI RNeasy columns (QIAGEN, Inc.). Contaminating DNA was eliminated using a DNA-Free kit (Ambion, Inc.). cDNA probes were generated with random primers (NSNSNSNSNS) annealed to total RNA (10 μg) and to five exogenous transcripts (ATCC 87482, 87483, 87484, 87485, and 87486; 130 pM). Random primers were annealed at 70°C for 10 min and 25°C for 10 min, followed by 37°C for 60 min, 42°C for 120 min, and 70°C for 10 min. Reactions contained 1,500 U Superscript reverse transcriptase II (Invitrogen Co.), 1× Superscript reverse transcriptase II buffer, 3 mM amino-allyl dUTP (Ambion, Inc.), 3 mM dTTP, 10 mM (each) dATP, dCTP, and dGTP (Invitrogen Co.), and 30 U SUPERase-In (Ambion, Inc.). Residual RNA was removed by alkaline treatment followed by neutralization, and cDNA was salt-ethanol precipitated at −70°C. Precipitated cDNA was resuspended in 0.2 M NaHCO3 (pH 9.0) and labeled with monoreactive Cy3/5 dyes in dimethyl sulfoxide (Amersham Pharmacia; as per manufacturer's instructions).
Labeled sample pairs were then combined accordingly and purified with a QIAquick PCR purification kit (QIAGEN, Inc.). The labeled cDNA sample was salt-ethanol precipitated and resuspended in hybridization buffer 2 (Ambion, Inc.). Custom in-house P. aeruginosa microarray slides (ArrayIT superamine slides) containing 400 to 600 cDNA probes (isolated as PCR amplicons) for 5,376 of the 5,570 P. aeruginosa genes were prepared for hybridization by washing two times for 5 min in 0.1% sodium dodecyl sulfate and five times for 1 min in water, boiling for 3 min in 95°C water, and air drying. Denatured sample was applied to a prepared microarray, overlaid with a Lifter-Slip coverslip (Electron Microscopy Sciences), and sealed in a hybridization chamber (Corning Inc.) for overnight hybridization at 45°C. Glass coverslips were removed by flotation in warm 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and slides were washed three times each for 5 min in 25 ml 0.5× SSC-0.5% sodium dodecyl sulfate and 25 ml 0.5× SSC and spun dry at 2,000 rpm for 5 min.
Microarrays were scanned on a ScanArray Express scanner (Perkin-Elmer) and images analyzed using ImaGene v5.0 (BioDiscovery, Inc.). The lowest 10 percent of signal for each subgrid was taken as background (R code developed by D. Hoffart and J. Brumm, Genome BC Array Facility). Output files were then compared and analyzed using R program scripts. All microarrays were normalized using the R package vsn (13). All genes in the 0.3× and 1× MIC of ciprofloxacin treatment group were compared to the untreated condition. To assess the statistical significance, two-sample t statistics and permutation P values were computed for each gene pair. Genes exhibiting permutation P values of ≤0.05 were considered differentially expressed. Data were also analyzed using GeneSpring v5.5 (Silicon Genetics) as a secondary analysis method. The changes (n-fold) presented herein are derived from GeneSpring v5.5 analysis and are shown only for those genes with statistically significant expression changes.
Microarray validation.
Relative real-time PCR was performed on the ABI Prism 7000 sequence detection system using SYBR green I dye chemistry (Applied Biosystems). Primers were designed using sequence from the Pseudomonas database (www.pseudomonas.com) and the ABI Primer Express program v2.0 in the default mode and are listed in Table 2. De novo cDNA was made from 0.3× MIC of ciprofloxacin-treated strain H103 RNA used in microarray target preparation. Data were analyzed using the ABI Sequence Detection software (ABI).
TABLE 2.
Primer sequences used in this study
| Primer | Sequence |
|---|---|
| PA0610_sense | 5′-TAGCACTCCGATTCCACGC-3′ |
| PA0610_antisense | 5′-CCGAAGATGCGGTAGACCA-3′ |
| PA0611_sense | 5′-AGCTTCAACCGCGAGGAATA-3′ |
| PA0611_antisense | 5′-CATGTCCTCCGGCGAGTACT-3′ |
| PA0621_sense | 5′-TTTCCCGTCAGCAACGTAGC-3′ |
| PA0621_antisense | 5′-GCTGACTATCCCGCCATCTC-3′ |
| PA0623_sense | 5′-CCGAGAAGCGCTGAATTTCT-3′ |
| PA0623_antisense | 5′-CCATTGAAAGCGCTCTGGTC-3′ |
| PA3617_sense | 5′-GTGAAGAACAAGGTTTCCCCG-3′ |
| PA3617_antisense | 5′-GAGGATCTGGAACTCGGCCT-3′ |
| PA3866_sense | 5′-CCACTTGTCGTGACCAGAGGA-3′ |
| PA3866_antisense | 5′-CATCGACCCAGGCTCGTAA-3′ |
| rplF_sense | 5′-AGGTTGCTGCCGAAATTCG-3′ |
| rplF_antisense | 5′-CTTGCCTTTGTAAGGCTCCG-3′ |
Luminescence changes from PA0620::lux and PA0641::lux were monitored over time using a SPECTRAFluorPlus luminometer (Tecan, San Jose, CA). Cells were grown as described above in LB broth supplemented with 0, 0.01, 0.03, or 0.1 μg/ml ciprofloxacin, 0, 50, 150, or 500 μg/ml novobiocin (Sigma-Aldrich Corp.), or 0, 0.2, 0.6, or 2 μg/ml ceftazidime (GlaxoSmithKline Beecham, Inc.). Triplicate samples were removed hourly for luminescence analysis and measurement of growth at an optical density of 600 nm; averaged luminescence values were corrected for growth. The assay was repeated in triplicate.
Pyocin/phage isolation and transmission electron microscopy.
Pyocin/phage-like particles were isolated and examined as previously described (29) from H103 and PA0620::lux cultures exposed to 1× the MIC of ciprofloxacin as described above for microarray experiments. Samples were placed on Formvar-coated grids, negatively stained with 1% phosphotungstate, and examined by transmission electron microscopy (Hitachi H7600 TEM) under standard operating conditions.
Microarray accession numbers.
RESULTS
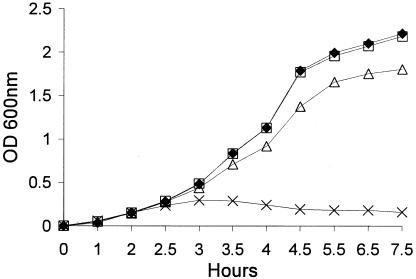
The pyocin/phage region is up-regulated in response to subinhibitory ciprofloxacin. The MIC of ciprofloxacin for P. aeruginosa PAO1 strain H103 (0.1 μg/ml [Table 3]) was comparable to the MIC for other sensitive Pseudomonas strains (5) and was used to calculate the 0.3× the MIC subinhibitory concentration. The growth rate of strain H103 was not found to be appreciably affected by subinhibitory or inhibitory concentrations of ciprofloxacin during a 2.5- to 3-h exposure; growth thereafter was severely limited by the MIC (Fig. 1). All subsequent experiments were conducted using cultures treated for 2.5 h.
TABLE 3.
MICs of different antimicrobials for various strains of P. aeruginosa grown in LB medium
| Strain | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|
| CIP | ENO | NOR | NAL | AMK | FEP | GAT | |
| H103 | 0.06 | 0.3 | 0.15 | 15.6 | 0.4 | 0.6 | 1.75 |
| prtR::lacZ | 0.06 | 0.3 | 0.15 | 31.25 | 0.4 | 0.6 | 0.875 |
| PA0613::lacZ | 0.5 | 1.6 | >2.5 | 250 | 0.4 | 0.6 | 0.875 |
| PA0620::lux | 0.5 | 1.6 | >2.5 | 250 | 0.4 | 1.25 | 0.875 |
| PA0621::lacZ | 0.5 | >5 | >2.5 | 250 | 0.2 | 0.3 | 0.875 |
| PA0641::lux | 0.5 | 1.6 | 1.25 | 125 | 0.4 | 1.25 | 1.75 |
| recA::phoA | 0.5 | 3.2 | >2.5 | 250 | 0.4 | 0.6 | 0.875 |
| PA3866::lux | 0.06 | 0.3 | 0.15 | 31.25 | 0.4 | 0.3 | ND |
Results shown are the mode of four independent experiments. CIP, ciprofloxacin; ENO, enofloxacin; NOR, norfloxacin; NAL, naldixic acid; AMK, amikacin; FEP, cefepime; GAENT, gentamicin; ND, not determined.
FIG. 1.
Growth curve for P. aeruginosa strain H103 in the presence or absence of ciprofloxacin. ⧫, untreated PAO1-H103; □, PAO1-H103 plus 0.01 μg/ml ciprofloxacin; ▵, PAO1-H103 plus 0.03 μg/ml ciprofloxacin; and ×, PAO1-H103 plus 0.1 μg/ml ciprofloxacin.
DNA microarray technology was then used to globally investigate the influence of subinhibitory (0.03 μg/ml = 0.3× MIC) and inhibitory (0.1 μg/ml = 1× MIC) concentrations of ciprofloxacin on the transcriptome of P. aeruginosa. Microarray analysis of P. aeruginosa strain H103 cells treated with 0.3× MIC of ciprofloxacin identified a total of 941 genes, approximately 15% of the known open reading frames (ORFs), as being significantly affected by the treatment (P ≤ 0.05) (a complete list of genes is available in the supplemental material). Of these, 554 were up-regulated and 387 down-regulated. Limitation of the data to twofold and fivefold expression differences produced more manageable lists of 243 genes and 16 genes, respectively. Microarray analysis of P. aeruginosa strain H103 cells treated with 1× MIC of ciprofloxacin identified a total of 1,230 genes that were significantly up- and down-regulated. A considerable number of ORFs on both of these lists fell into either the “hypothetical gene” or “putative enzyme” functional categories (27) (52 and 579 ORFs for 0.3× and 1× MIC treatments, respectively), illustrating that a large proportion of genes responsive to ciprofloxacin remain to be fully characterized. Moreover, the expression changes for many of these ORFs tended to be fairly large in comparison to the expression changes observed for the annotated genes, highlighting the potential importance of these uncharacterized ORFs in the cellular response of P. aeruginosa to ciprofloxacin.
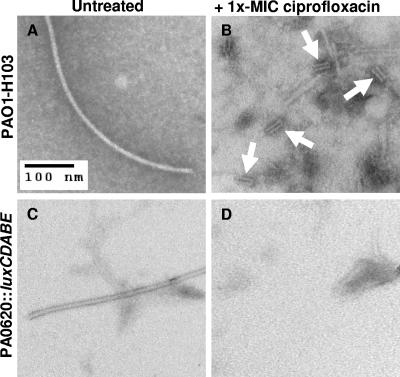
Most striking among the responsive genes was the region between PA0613 and PA0648, where virtually every ORF was increased and where the expression level for ORFs in this region became more pronounced with the increasing concentration of ciprofloxacin (Table 4). PA0616 to PA0648 codes for the R2 and F2 pyocins, which share homology with various bacteriophage-related proteins (21). Both pyocins are known to resemble phage tails and appear to have evolved as bacteriocins rather than being simple defective phages (21). Consistent with this and the observed expression trends, phage tail particles were noted by electron microscopy in the supernatants of wild-type P. aeruginosa cells treated with 1× MIC of ciprofloxacin but not in the supernatants of untreated wild-type or ciprofloxacin-treated strain PA0620::luxCDABE containing a polar mutation in this R2 pyocin ORF (Fig. 2). The filamentous particles observed in Fig. 2A and C likely represent cleaved flagella, as observed by Higerd et al. (10).
TABLE 4.
Pyocin/phage operon and related genes induced by 0.3× and 1× MIC of ciprofloxacina
| ORF | Gene nameb | Change (n-fold) with 0.3× MIC | Change (n-fold) with 1× MIC | Description |
|---|---|---|---|---|
| PA0610 | prtN | 2.0 | 6.8 | Transcriptional activator |
| PA0611 | prtR | 1.3 | 3.0 | Transcriptional repressor |
| PA0612 | —c | 8.5 | Homolog of Zn2+ finger protein | |
| PA0613 | 4.0 | 19.3 | Conserved hypothetical | |
| PA0614 | Hol | 3.3 | 15.3 | Holin |
| PA0615 | 1.7 | 7.2 | Conserved hypothetical | |
| PA0616 | VR2 | 2.9 | 13.0 | Homologous to baseplate assembly protein V |
| PA0617 | WR2 | 3.0 | 13.9 | Homologous to baseplate assembly protein W |
| PA0618 | JR2 | 2.1 | 10.0 | Homologous to baseplate assembly protein J |
| PA0619 | IR2 | 3.2 | 14.7 | Homologous to tail protein I |
| PA0620 | HR2 | 2.7 | 12.1 | Homologous to tail fiber protein H |
| PA0621 | 4.3 | 16.7 | Homologous to tail fiber assembly protein | |
| PA0622 | FIR2 | 2.4 | 10.3 | Homologous to contractile sheath protein FI |
| PA0623 | FIIR2 | 2.9 | 12.8 | Homologous to tail tube protein FII |
| PA0624 | 2.7 | 12.8 | Conserved hypothetical | |
| PA0625 | 2.5 | 14.1 | Homologous to tail length determination protein | |
| PA0626 | UR2 | 1.8 | 9.8 | Homologous to tail formation protein U |
| PA0627 | XR2 | 2.3 | 10.8 | Homologous to tail protein X |
| PA0628 | DR2 | 1.8 | 10.0 | Homologous to tail formation protein D |
| PA0629 | Lys | 2.2 | 8.7 | Lytic protein; homology to predicted chitinase |
| PA0630 | 1.6 | 10.6 | Hypothetical protein | |
| PA0631 | 2.4 | 16.2 | Unique hypothetical protein | |
| PA0632 | 3.4 | 21.2 | Unique hypothetical protein | |
| PA0633 | VF2 | 4.3 | 16.4 | Homologous to major tail protein V |
| PA0634 | 3.2 | 15.8 | Unique hypothetical protein | |
| PA0635 | 2.0 | 15.9 | Conserved hypothetical protein | |
| PA0636 | HF2 | 2.1 | 10.0 | Homologous to tail length determination protein H |
| PA0637 | MF2 | 2.7 | 11.0 | Homologous to tail fiber protein M |
| PA0638 | LF2 | — | 8.5 | Homologous to tail fiber protein L |
| PA0639 | KF2 | 3.0 | 15.9 | Homologous to tail assembly protein K |
| PA0640 | IF2 | 3.1 | 16.4 | Homologous to tail assembly protein I |
| PA0641 | JF2 | 2.4 | 9.8 | Homologous to tail fiber protein J |
| PA0642 | 2.7 | 12.9 | Hypothetical protein | |
| PA0643 | 1.9 | 8.9 | Homologous to tail fiber domain protein | |
| PA0644 | 4.0 | 18.0 | Hypothetical protein | |
| PA0645 | 1.8 | 15.8 | Hypothetical protein | |
| PA0646 | 3.1 | 13.4 | Homologous to putative tail fiber protein | |
| PA0647 | 3.8 | 23.0 | Conserved hypothetical protein | |
| PA0648 | — | 8.2 | Conserved hypothetical protein | |
| PA0985 | pys5 | 4.5 | 18.1 | Pyocin S5 |
| PA1150 | pys2 | — | 5.4 | Pyocin S2 |
| PA3617 | recA | 2.8 | 4.6 | Recombinase for DNA recombination and repair |
| PA3866 | pys4 | 13.9 | 51.3 | Pyocin S4 |
Only genes identified as being affected by subinhibitory (0.3 μg/ml) or inhibitory (1 μg/ml) ciprofloxacin relative to untreated strain H103 and that were of interest in this study are included. A list of all genes identified as being affected by this concentration of ciprofloxacin and exhibiting a statistically significant change (P ≤ 0.05) is available in the supplemental material. Genes are identified by ORF designation, gene name or alternative gene name, and homology description based on the Pseudomonas genome project (www.pseudomonas.com).
Genes were named as per Table 1 of Nakayama et al. (21) and reflect the homology observed by these authors to phages P2 and λ.
—, no significant change in expression.
FIG. 2.
Electron micrographs of supernatants from P. aeruginosa cells that were untreated or treated with 1× MIC of ciprofloxacin. (A) Untreated strain H103 cells. (B) 1× MIC of ciprofloxacin-treated strain H103 cells. Arrows point to the presence of pyocin/phage tail structures. (C) Untreated strain PA0620::luxCDABE cells. (D) 1× MIC of ciprofloxacin-treated PA0620::luxCDABE cells showing absence of R-type pyocin tail structures.
Regulation of this region, as well as that of other P. aeruginosa pyocins (i.e., PA0985, PA1150, and PA3866), has been well documented (19) and involves RecA (PA3617) control of the repressor protein PrtR (PA0611), allowing transcription to proceed through interaction of the activator PrtN (PA0610) with the P-box motif (25). Both the regulatory network ORFs and the other pyocins in P. aeruginosa were also up-regulated by subinhibitory and inhibitory ciprofloxacin (Table 4), reinforcing the idea that ciprofloxacin induces the expression of pyocins. These observations were consistent with the responsiveness of pyocins and lysogenic phage to mutagenic and/or DNA-damaging events (20), given that ciprofloxacin is known to interfere with DNA replication and thus have mutagenic activity (18, 20).
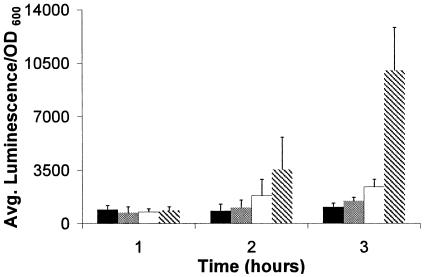
The array data were validated by analyzing the expression of several R2/F2 pyocin ORFs by relative real-time PCR. Expression trends were found to be similar to those observed for the microarray data (Table 5). Expression changes were also confirmed by measuring over a 3-hour time course in the presence of 0.1×, 0.3×, and 1× MIC of ciprofloxacin the luminescence from mini-Tn5luxCDABE transcriptional fusions inserted in the PA0620 (Fig. 3) and PA0641 (data not shown) genes. Similar up-regulation trends to the array data were noted for the DNA gyrase inhibitors ciprofloxacin (Fig. 3) and novobiocin but not for the β-lactam ceftazidime, highlighting the responsiveness of the R2/F2 pyocin region to DNA gyrase inhibition.
TABLE 5.
Comparison of expression changes for various pyocin/phage-related open reading frames as analyzed by relative real-time PCR and custom P. aeruginosa microarray
| ORF | Gene Name | Change (n-fold) in PAO1 strain H103 at 0.3× MIC of ciprofloxacin
|
|
|---|---|---|---|
| Custom microarray | Real-time PCR | ||
| PA0610 | prtN | 2.0 | 10.0 |
| PA0611 | prtR | 1.3 | 11.4 |
| PA0621 | 4.3 | 3.0 | |
| PA0623 | FIIR2 | 2.9 | 3.7 |
| PA3617 | recA | 2.8 | 3.9 |
| PA3866 | pys3 | 13.9 | 22.5 |
FIG. 3.
Induction of PA0620::luxCDABE fusions in LB media alone (black bars), with 0.1× MIC of ciprofloxacin (0.01 μg/ml; grey bars), with 0.3× MIC of ciprofloxacin (0.03 μg/ml; white bars), or with 1× MIC of ciprofloxacin (0.1 μg/ml; patterned bars).
Mutants in pyocin/phage region have higher fluoroquinolone MICs.
To determine whether these striking expression changes in the pyocin/phage region in response to ciprofloxacin were relevant to susceptibility/resistance, mutants deficient in a number of these genes (Table 1) were tested for antibiotic sensitivity. MICs of all four fluoroquinolones examined were generally ≥8-fold higher for PA0613::ISlacZ, PA0620::lux, PA0621::ISlacZ, and PA0641::lux mutants compared to the wild-type P. aeruginosa strain H103 (Table 3). No significant changes in the MICs were observed for the control antibiotics amikacin, cefepime, or gentamicin. A recA::ISlacZ mutant also exhibited eightfold-higher MICs for all four fluoroquinolones, while a mutant in prtR, the transcriptional repressor, showed no difference in MICs relative to wild-type strain H103 (Table 3). Together, these surprising results indicate that expression of the R2/F2 pyocin region in the wild-type strains determines (i.e., increases) the intrinsic susceptibility of P. aeruginosa to ciprofloxacin and other fluoroquinolones at the MIC.
DISCUSSION
To better address the rise in bacterial resistance and develop novel antimicrobials to combat these infections, we must first better understand the response of an organism to antibiotic challenge. While it has long been believed that the target and/or mechanism of action of certain antimicrobials, like ciprofloxacin, is well understood, recent microarray work on transcriptional responses to drugs with the same proposed mechanism of action stands in stark contradiction to this notion. For example, although novobiocin and ciprofloxacin both target DNA gyrase, distinct expression patterns in response to these two agents have been observed in H. influenzae and Bacillus subtilis (7, 14). Furthermore, the expression signatures of other DNA-damaging compounds, namely, hydrogen peroxide and doxorubicin, but not ethidium bromide, have been shown to be closely related to the expression profiles of quinolone topoisomerase inhibitors (14). Examination of the transcriptional responses of P. aeruginosa to subinhibitory and inhibitory ciprofloxacin likewise showed an unanticipated finding, in that increased expression of the R2/F2 pyocin region in response to treatment with ciprofloxacin at or below the MIC seems to play a critical role in mediating susceptibility to ciprofloxacin and other fluoroquinolones. Thus, in addition to its ability to interact with DNA topoisomerases, ciprofloxacin also induces the expression of this susceptibility determinant. No significant changes in the genes encoding the target DNA gyrase were observed after treatment with 1× MIC of ciprofloxacin, although such changes were not anticipated, since Gmuender et al. (7) observed changes in DNA gyrase target genes only with 10× MIC of ciprofloxacin.
The work of Nakayama et al. (21) provides a possible explanation for how the R2/F2 pyocins may encode a fluoroquinolone susceptibility determinant. In their analysis of the R2/F2 pyocin region, they presented evidence of the existence of a phage lytic system. PA0614 exhibited a hydrophobicity profile similar to holins of phage P2, φCTX, and λ phage. Holins mediate the translocation of lytic enzymes to the outer membrane. Analysis of PA0629 found weak but significant similarity to chitinases, enzymes which have substrate specificities similar to those of lysozymes which degrade bacterial peptidoglycan, giving rise to the suggestion that PA0629 may be a lytic enzyme required for pyocin secretion (21). Consistent with this proposal, expression of PA0614 and/or PA0629 from an inducible plasmid was shown to result in cell lysis in Escherichia coli and P. aeruginosa (21). Given this proposed phage lytic system in the R2/F2 pyocin region, it seems plausible that the lethality associated with ciprofloxacin induction of the pyocin region may be due to induction of this lysis system. Indeed, growth of P. aeruginosa PAO1-H103 was inhibited by 1× MIC of ciprofloxacin (Fig. 1), and Gram stains of these cultures showed diminished numbers of cells over exposure time (data not shown). Correspondingly, individual mutants in both the R2 and F2 regions (i.e., mutants in genes PA0613, PA0620, PA0621, and PA0641) exhibited resistance to ciprofloxacin (Table 3). These results highlight the importance of cell lysis in R2/F2 pyocin-mediated susceptibility to ciprofloxacin. Conversely, resistance to ciprofloxacin was not observed for mutants in another pyocin gene, PA3866 (Table 3), even though it was even more strongly induced by subinhibitory and inhibitory concentrations of ciprofloxacin, indicating that this phenomenon is specific to the R2/F2 pyocins.
Genomic analysis of many clinical and environmental isolates of P. aeruginosa has demonstrated significant mosaicism in the R2/F2 pyocin region (4, 28). Interestingly, all strains studied contained either the R2 pyocin or the F2 pyocin regions, as assessed by microarray analysis (4, 28); the putative holin gene PA0614 and lysis gene PA0629, however, were found to be conserved across all eight clinical isolates studied. Recent work on colicins (E. coli equivalent of pyocins) and their role in biodiversity has identified them as having the ability to promote microbial diversity in static or localized environments, in a manner similar to the game rock-paper-scissors (16, 17). In this model, strains that produce colicin kill sensitive strains, which outcompete resistant strains, which, in turn, outcompete colicin-producing strains on the basis of growth rate advantages. However, in well-mixed or large spatial environments, the resistant strain will predominate over time (16). While this work does not demonstrate genomic loss of the pyocin region, it does indicate that there is a selective pressure favoring colicin-resistant strains. Similarly, progressive loss of pyocin production in cystic fibrosis clinical isolates has been observed (11, 24) and occurs while patients are under selective pressure, as antimicrobial therapy is applied in an attempt to control their lung disease. It is intriguing to speculate that parts or all of the pyocin region can be deleted in P. aeruginosa clinical isolates due to strain competition caused by enhanced survival of the pyocin-resistant strain and that this deletion is further advantageous with respect to the fluoroquinolone resistance profile of the organism. Further studies to investigate this prospect are required, since their findings have important implications in the role subinhibitory antimicrobials play in the development of resistance in P. aeruginosa.
Supplementary Material
Acknowledgments
Funding for this research work was generously supported by the Canadian Cystic Fibrosis Foundation (CCFF), the Canadian Institutes of Health Research, and the Functional Pathogenomics of Mucosal Immunity program funded through Genome Prairie and Genome BC, with additional contributions from Inimex Pharmaceuticals. M.D.B. was supported by the CCFF and NSERC, and R.E.W.H. is the recipient of a Canada Research Chair.
Pathogenesis, Inc. (subsequently acquired by Chiron), and particularly Ken Stover and Silvija Coulter, are gratefully acknowledged for their assistance in constructing the DNA probes for the custom DNA microarray. Colleen Nelson and members of the Gene Array Facility at the Prostate Cancer Centre, VGH, are also acknowledged for their excellent technical support during the microarray printing process. We thank Jenny Bryan and Dana Aeschlimann in the Department of Statistics at UBC for their statistical expertise and generous contributions to this project.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Amsterdam, D. 1991. Susceptibility testing of antimicrobials in liquid media, p. 72-78. In V. Lorian (ed.), Antibiotics in laboratory medicine. Williams & Wilkins, Baltimore, Md.
- 2.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 3.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 4.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 5.Fung-Tomc, J., B. Kolek, and D. P. Bonner. 1993. Ciprofloxacin-induced, low-level resistance to structurally unrelated antibiotics in Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 7.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, R. E., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updates 3:247-255. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. E. W. 1997. The bacterial outer membrane as a drug barrier. Trends Microbiol. 5:37-42. [DOI] [PubMed] [Google Scholar]
- 10.Higerd, T. B., C. A. Baechler, and R. S. Berk. 1967. In vitro and in vivo characterization of pyocin. J. Bacteriol. 93:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holloway, B. W., H. Rossiter, D. Burgess, and J. Dodge. 1973. Aeruginocin tolerant mutants of Pseudomonas aeruginosa. Genet. Res. 22:239-253. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, D. C. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist. Updates 2:38-55. [DOI] [PubMed] [Google Scholar]
- 13.Huber, W., A. Von Heydebreck, H. Sultmann, A. Poustka, and M. Vingron. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18:S96-S104. [DOI] [PubMed] [Google Scholar]
- 14.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. (First published 14 November 2003; 10.1073/pnas.2036282100.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr, B., M. A. Riley, M. W. Feldman, and B. J. Bohannan. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 17.Kirkup, B. C., and M. A. Riley. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412-414. [DOI] [PubMed] [Google Scholar]
- 18.Mamber, S. W., B. Kolek, K. W. Brookshire, D. P. Bonner, and J. Fung-Tomc. 1993. Activity of quinolones in the Ames Salmonella TA102 mutagenicity test and other bacterial genotoxicity assays. Antimicrob. Agents Chemother. 37:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsui, H., Y. Sano, H. Ishihara, and T. Shinomiya. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 175:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 22.Ng, W. L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen, S. S., N. Hoiby, F. Espersen, and C. Koch. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romling, U., B. Fiedler, J. Bosshammer, D. Grothues, J. Greipel, H. von der Hardt, and B. Tummler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 25.Sano, Y., H. Matsui, M. Kobayashi, and M. Kageyama. 1993. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 175:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw, K. J., and B. J. Morrow. 2003. Transcriptional profiling and drug discovery. Curr. Opin. Pharmacol. 3:508-512. [DOI] [PubMed] [Google Scholar]
- 27.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 28.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. (First published 18 June 2003; 10.1073/pnas.0832438100.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto, K. R., B. M. Alberts, R. Benzinger, L. Lawhorne, and G. Treiber. 1970. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40:734-744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.