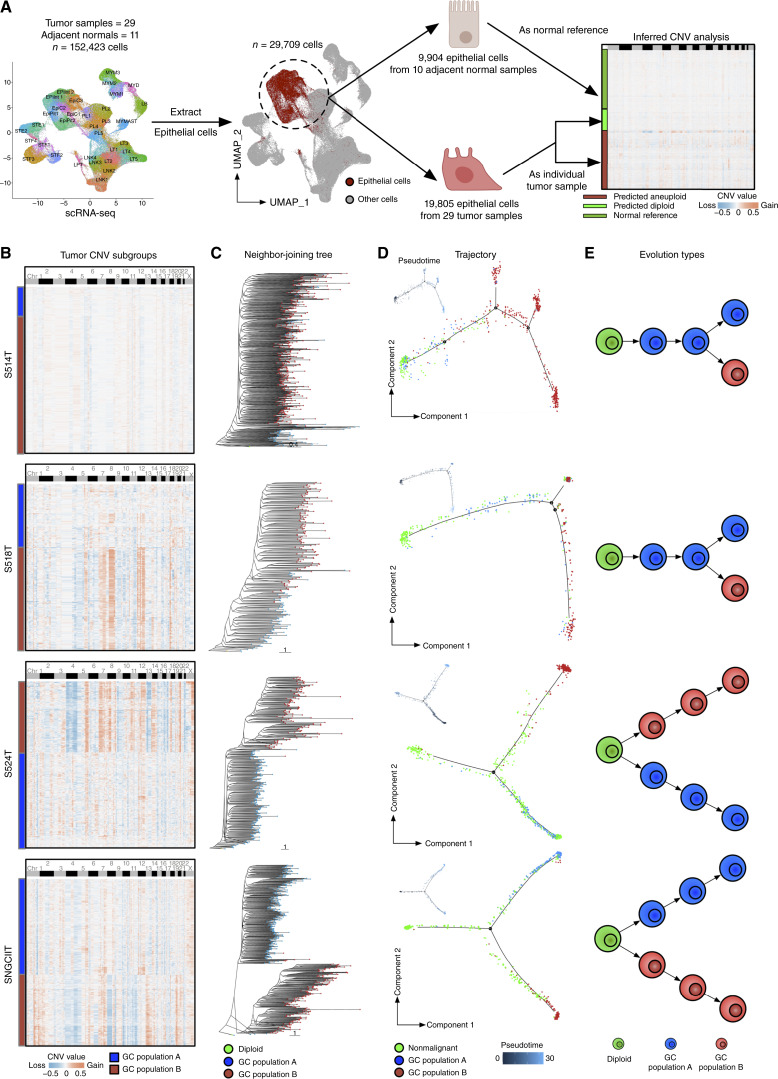
Figure 3.
Single-cell resolved evolutionary trajectories in gastric cancer. A, Workflow depiction for sCNA inference from scRNA-seq single-cell data. Adjacent normal epithelial cells from 10 adjacent normal samples were used as references (one normal sample was filtered out due to the lack of a matched tumor sample), and tumor epithelial cells from 29 patients were classified as diploid and aneuploid. B, Heatmap presenting inferred sCNA values for tumor subgroups within single-cell data for samples S514T, S518T, S524T, and SNGCIIT (from top to bottom). Rows represent individual cells, whereas columns correspond to genomic bin positions of 220 kb each. Color gradations indicate varying states of sCNA, with subgroups determined through unsupervised clustering based on Euclidean distances. C, Neighbor-joining tree constructed on the inferred sCNA values, with the tree rerooted to a diploid state for reference. Each dot on a tree branch represents a single cell, color-coded according to subgroup labels identified in B. D, Main panels show trajectory plots, whereas the top left panels show pseudotime graphics for nonmalignant cells and tumor cells from identified sCNA subgroups. Each dot represents a cell, color-coded by cell type. Solid lines in the trajectory plot are derived from single-cell expression data, indicating the developmental path of cells. Pseudotime analysis, with zero time points anchored to selected nonmalignant cells, use color gradations to represent estimated pseudotimes for each cell. E, The illustrated figure conceptualizes the two identified tumor evolution patterns in gastric cancer. Cells in green are depicted as diploids, whereas cells in blue and red symbolize two distinct tumor populations, each characterized by their sCNA patterns. GC, gastric cancer.