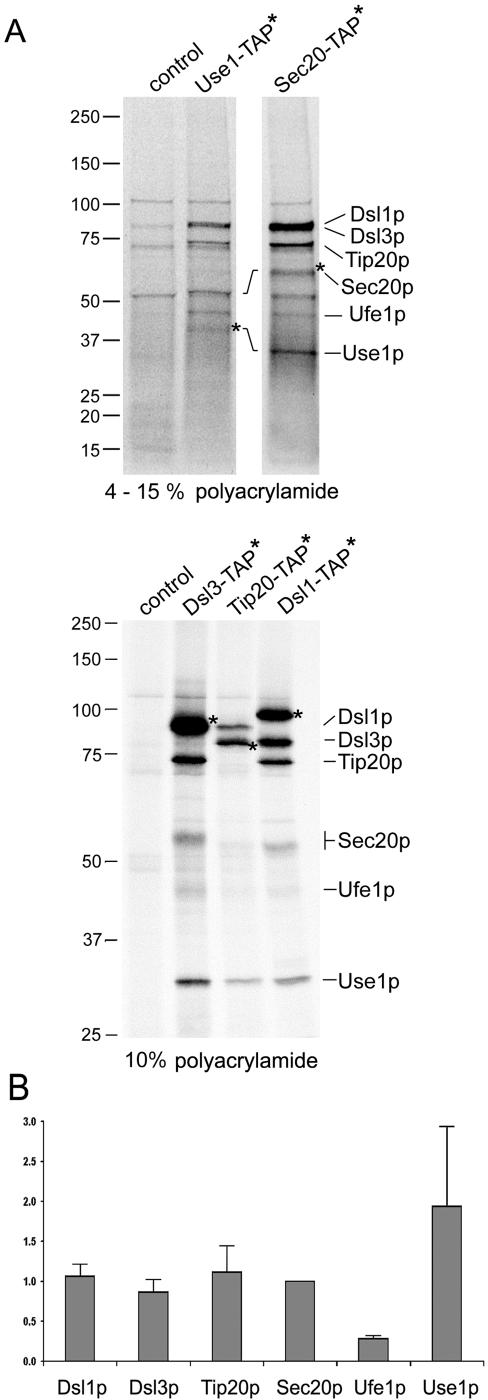
Figure 6.
Tandem affinity purification of the Dsl1p complex and associated SNARE proteins. The composition of the Dsl1p complex was analyzed by applying a modified tandem affinity purification protocol (Rigaut et al., 1999) as described in Materials and Methods. Cells expressing the TAP-tagged SNARE proteins Use1p and Sec20p as well as the Dsl1p complex subunits Dsl1p, Dsl3p, and Tip20p were labeled with 35S-methionine and -cysteine to facilitate the detection of purified proteins. The wild-type strain BY4741 was also analyzed to visualize proteins that bind unspecifically to IgG Sepharose and protein A Sepharose carrying anti-TAP antibodies. Proteins were identified by an additional immunoprecipitation. Before the reprecipitation, these samples had been incubated at 95°C for 5 min to disassemble protein complexes (Supplementary Figure S4A). Samples were loaded either on a 4–15% gradient SDS polyacrylamide gel (Bio-Rad; top panel) to distinguish small proteins or on a one-percentage 10% gel to better resolve the larger proteins (bottom panel). For comparison, Supplementary Figure S4B shows a one-percentage gel with a preparation using a Sec20-TAP expressing strain and a 4–15% gradient gel with samples from Dsl3-TAP, Tip20-TAP, and Dsl1-TAP expressing strains. The migration of molecular-weight markers is indicated on the left. (B) Phosphoimager pictures from two independent Sec20-TAP preparations as well as one Dsl3-TAP, Tip20-TAP, and Dsl1-TAP preparation were quantified (AIDA Biopackage, Raytest). Data were corrected for different contents of methionine and cysteine of the proteins. The intensity values for each protein were normalized to the number of methionines and cysteines in Sec20p (minus the initiator methionine). Sec20p with eight methionines and cysteines was assigned a normalization factor of 1 because it was discernable as a single band in all preparations.